Abstract
About 160 species are classified within the Viburnum genus and many of these are cultivated for horticultural purposes. The vast dispersal of Viburnum makes the genus a useful model for studying evolutionary history and inferring how species expanded into their current distributions. Simple sequence repeat (SSR) markers were previously developed for five Viburnum species that were classified within the four major clades (Laminotinus, Crenotinus, Valvatotinus, and Porphyrotinus). The ability of some of these markers to cross-amplify in Viburnum species has been scantly evaluated, but there has not been any genus-wide assessment for the markers. We evaluated a collection of 49 SSR markers for the ability to cross-amplify in 224 samples, including 46 Viburnum species, representing all 16 subclades, and five additional species in the Viburnaceae and Caprifoliaceae. A subset of 14 potentially comprehensive markers for Viburnum species was identified and evaluated for the ability to detect polymorphisms in species outside of their respective clades. The 49 markers had overall amplification success in 52% of the samples, including a 60% success rate within the Viburnum genus and 14% in other genera. The comprehensive marker set amplified alleles in 74% of all samples tested, including 85% of Viburnum samples and 19% of outgroup samples. To the best of our knowledge, this is the first comprehensive set of markers able to characterize species across an entire genus. This set of markers can be used to assess the genetic diversity and population structure of most Viburnum species and closely allied species.
Similar content being viewed by others
Introduction
Viburnum L. was formerly classified within the Caprifoliaceae (honeysuckle family) alongside Lonicera L. and Weigela Thunb. Viburnum is a large genus comprised of approximately 160 species1, now classified in the Viburnaceae (formerly Adoxaceae)2,3 with Adoxa L., Sambucus L., and a few other genera. Species within Viburnum are native to temperate and subtropical regions of the Northern Hemisphere, and the range extends into the mountainous regions of Southeastern Asia and South America. The somatic chromosome numbers of Viburnum species range from 18 to 724,5 and in ploidy level from diploid to octaploid. Not all species have been equally studied, but most of the species are 2n = 2x = 184,5,6. Studied Viburnum genome sizes are classified as small to intermediate when compared to plants overall and have 2C values ranging from 4.29 to 24.23 Gbp7,8,9,10,11. However, the genome sizes are larger than some other woody genera, such as Cornus with 2C values ranging from 1.89 to 6.66 Gbp7,12,13,14 and Pyrus ranging from 1.13 to 1.27 Gbp10,15,16.
The genus Viburnum includes many species of shrubs to small trees with year-round ornamental qualities, which are grown for their fragrant flowers and attractive foliage. There are 70 species and interspecific hybrids in cultivation17 that generated $23.2 million in wholesale and retail sales in 201918. This well-sampled, geographically widespread, and abundant genus also provides a model for addressing evolution, biogeography, phylogenetics, and ecology-related questions, and has been the topic of many investigations1,19,20,21,22.
Much progress has been made in the phylogenetic classification of this genus6,21,23,24,25,26, but knowledge gaps persist in the taxonomy that if resolved, would provide better insight into the evolution, biogeography, and phylogenetics of the genus. DNA-barcoding was implemented but had a low success rate due to low sequence variability25. Restriction-site-associated DNA sequencing (RADseq) was applied to the V. nudum species complex in North America and was successful in identifying three independent lineages in support of three separate species27. However, this next generation sequencing (NGS) approach may not be feasible for every species in this genus because of the relatively large costs associated with NGS in addition to the large genome sizes and ploidy levels of some Viburnum spp.
Four sets of microsatellite markers, also known as simple sequence repeat (SSR) markers, were developed from species within the four major clades of Viburnum22 including Laminotinus (Succotinus subclade)28, Valvatotinus (Lentago subclade)29, Porphyrotinus (Oreinotinus subclade)30, and Crenotinus (Solenotinus subclade)31 (Fig. 1). SSRs consist of short nucleotide motifs that are tandemly repeated. The nucleotide motifs can be between one and five base pairs (bp) long and are ubiquitous throughout the genome32. These regions of the genome can accumulate mutations faster than others predominantly due to polymerase slippage during DNA replication and are flanked by relatively conserved genetic sequences, thus allowing for the design of PCR primers to consistently amplify a given locus33,34. The resulting amplicons can then be sized or sequenced to determine the alleles in an individual. The co-dominant nature, hypervariability, reproducibility, and PCR applicability make these genetic markers a popular option for population analyses35, species and cultivar delimitation36, and breeding37. Furthermore, SSR markers often transfer to closely related species and genera, and only require small amounts of low-quality DNA for successful amplification. A cost analysis was performed in 2020 on SSR markers used in conjunction with the QIAxcel capillary electrophoresis system and determined the cost per sample per locus to be less than $1238, making this a very affordable option. The combination of high polymorphism, low cost, and low-quality DNA requirements make SSRs an optimal tool for the vast Viburnum genus.
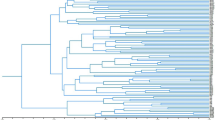
Phylogenetic tree of Viburnum species included in this study. This tree is a trimmed version of the one published in Landis et al.1 based on RAD-seq data. Highlighted nodes are species in which simple sequence repeat (SSR) markers were developed. Subclades are horizontal and major clades are vertical.
Using SSR markers developed in other species to study related species is a common practice and has been demonstrated in various ornamental taxa such as Cornus (dogwood)39 and Cercis (redbud)40 as well as food crops such as Glycine (soybean)33, Prunus (stonefruit)41, and Foeniculum (fennel)42. A meta-analysis was completed on studies reporting cross-species amplification success in plants, fungi, and animals, and determined for eudicots overall. For eudicots the percentage of markers that were able to cross-amplify in species within genera was almost 80% and almost 60% among genera, whereas the percentage of markers that were actually polymorphic was almost 60% within genera and 10% among genera43. Some of the Viburnum markers were demonstrated to cross-amplify in their initial development publications, and a preliminary cross-amplification analysis was performed as part of Dean’s dissertation44. Additionally, the markers developed from V. dilatatum were used to amplify alleles in V. opulus, V. trilobum, and V. sargentii45, but no large-scale analysis has been published with any of the four sets.
The development of unique marker sets for each of the approximately 160 Viburnum species1 would be cost- and time-prohibitive. Therefore, the overarching goal of the study was to develop a set of comprehensive markers that could be applied to studies of genetic diversity, population genetics, and potentially phylogenetics of all the species within Viburnum as well as some closely allied genera in the Viburnaceae and Caprifoliaceae. We hypothesized this comprehensive marker set could be identified from the previously published markers. To test this the following aims were completed: 1) evaluate the cross-amplification of the 49 previously published markers in 46 Viburnum species and five species in the Viburnaceae and Caprifoliaceae; 2) compile a set of comprehensive markers for the genus, and 3) demonstrate the ability of the comprehensive marker set to detect polymorphisms in species outside of the subclades from which they were developed.
Materials and methods
Plant materials and gDNA extraction
Leaf samples were donated by arboreta, herbaria, and public universities across the U.S. except for the few species found locally in East Tennessee, U.S. (Supplementary Table 1). The use of samples in the present study complied with international, national and/or institutional guidelines. Local samples were collected and identified by Robert Trigiano and William Klingeman from public areas, which did not require permission to collect (see Supplementary Table 1 for additional information). The goal was to obtain three independent samples for each species and three species from each of the 16 subclades, following the classification scheme proposed in Clement et al.22 (i.e., Lentago, Punctata, Euviburnum, Pseudotinus, Urceolata, Solenotinus, Lutescentia, Tinus, Sambucina, Coriacea, Succotinus, Lobata, Opulus, Mollotinus, Dentata, and Oreinotinus). For the purposes of this study, these 16 subclades will be referred to as subclades and Valvatotinus, Crenotinus, Porphyrotinus, and Laminotinus will be referred to as major clades, despite there being higher classifications than these four. For additional information about the formal phylogenetic definitions, see Clement et al.22. Due to limited sampling of some subclades, it was not possible to meet the sampling goal for every subclade, but 46 Viburnum species and five closely related species in the Viburnaceae and Caprifoliaceae were obtained for a total of 224 samples (Supplementary Table 1). The Viburnaceae and Caprifoliaceae are both classified within Dipsacales, making Caprifoliaceae species ideal candidates for outgroup samples. For visualization of relatedness within Viburnum, the phylogenetic tree from Landis et al.1 was trimmed with the R packages ape (version 5.6–1) and ggtree (version 3.2.1) to only include the Viburnum species represented in this study (Fig. 1).
Genomic DNA (gDNA) isolations followed the same protocol as described in Hamm et al.31. Leaf samples were frozen in liquid nitrogen before homogenization using a Beadmill 24 (Fishers Scientific, Pittsburgh, Pennsylvania, U.S.). All leaf samples from arboreta were air-dried between pieces of newspaper before freezing. gDNA was extracted from leaf samples using the Omega E.Z.N.A. Plant DNA Kit (Omega Bio-tek Inc., Norcross, Georgia, U.S.), following the manufacturer’s protocol except that 2% mass/volume polyvinylpyrrolidone (PVP) (Fisher BioReagents, Waltham, Massachusetts, U.S.) was added to the P1 Buffer and the incubation time at 65 °C was increased from 10 to 30 min. DNA from herbaria samples dating back to 1932 was successfully extracted with this kit. A CTAB protocol46 was used with a few herbaria samples because of the paucity of leaf material. The quality of the extracted gDNA was assessed with a NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.).
SSR primers and genotyping conditions
A total of 49 primer pairs from four previous studies that were developed from the four major clades of Viburnum were used in this study. Eleven markers were developed from V. dilatatum in Laminotinus28, seven from V. rufidulum in Valvatotinus29, 16 from V. triphyllum and V. pichinchense in Porphyrotinus30, and 15 from V. farreri in Crenotinus31 (Supplementary Table 2). All SSR markers were single locus and polymorphic in the species from which they were obtained and included simple-perfect and compound-imperfect motifs. For simplicity, the markers developed from V. triphyllum and V. pichinchense were renamed Vore01-16 (Vore = Viburnum Oreinotinus subclade) in the same order found in the published table (original names in parentheses in Supplementary Table 2)30.
PCR was completed with all 49 primer pairs and 224 samples. A reaction volume of 10 µL, consisting of 5 µL of 2X Accustart II PCR SuperMix (Quantabio, Qiagen Beverly, Inc., Beverly, Massachusetts, U.S.), 3 µL autoclaved water, 1 µL of a mixture of 5 µM forward and primers, and 1 µL of 2 ng/µL gDNA was used. A single PCR thermal profile was used with all samples and markers and was as follows: 3 min of initial denaturation at 94 °C, 10 touchdown47 cycles (94 °C for 40 s, 60 °C–0.5 °C/cycle for 40 s, and 72 °C for 45 s) and 30 cycles (94 °C for 40 s, 55 °C for 40 s, 72 °C for 45 s), and a final extension at 72 °C for 4 min. This thermal profile was selected as a compromise among all the varying protocols from the initial development publications. PCR products were visualized with capillary electrophoresis (QIAxcel Advanced Electrophoresis System; Qiagen) and analyzed using a 25- to- 500 base pair (bp) DNA size marker (Qiagen) and an internal 15/600 bp alignment marker (Qiagen). Due to the wide variety of species and varying genetic distances among the samples, four positive controls were included in every 96-well PCR plate. The positive control samples were all from freshly collected leaves of species that were used in the original marker development (V. dentatum, V. dilatatum, V. farreri, and V. rufidulum). A negative control of sterile water was also included in every plate. Allele sizes were determined using QIAxcel ScreenGel version 1.6.0.10.
Scoring of amplification and dataset analysis
Only peaks greater than 0.1 Relative Fluorescent Units (RFUs) were considered amplified alleles. For diploid species, only the two strongest peaks were scored. For polyploid species, the number of peaks considered equaled the ploidy level. For example, a total of eight peaks were considered for the octaploid species, V. bracteatum. The highest known ploidy in this genus is octaploid, therefore for any species with an unknown ploidy level, a maximum of eight peaks was considered as well. Any amplicon greater than 600 bp was not sized because it was outside the 600 bp maximum of the alignment marker. A reaction was considered within the expected bp range and a successful cross-amplification if the amplicon size was within approximately 50 bp of the expected allele size from the original characterized species. Amplicons outside of the expected range were assumed to be from non-target loci. The 50 bp cutoff was selected because most of the strong peaks with limited noise were within 50 bp of the expected range. If an amplicon was outside of that range, it was recorded as an asterisk in the datasheet. If a reaction resulted in no amplification, the PCR was attempted again for all samples except L. japonica and maackii. For these outgroup species, if there was no amplification the marker was not repeated, however, if there was amplification with a single sample of these two species, all L. japonica and maackii samples were repeated. From QIAxcel, the dataset was then imported into Jupyter Notebook for analysis and visualization with custom code (https://github.com/trinityhamm/Viburnum_Cross_Amp/blob/main/Viburnum_Cross_Amp_final.ipynb).
Validation: comprehensive marker set
Once cross-amplification was completed with all 49 markers, a comprehensive marker set was identified. The 14 markers identified displayed high amplification rates across most subclades. To demonstrate that these markers could characterize species outside of the subclade they were developed in, a subset of the data was analyzed more closely. More than three leaf samples were obtained for V. carlesii (n = 14), V. opulus (n = 18), V. plicatum (n = 15), and V. tinus (n = 9), which are all members of subclades where markers were not developed from. The amplification rates and observed heterozygosity were then calculated for each of the 14 comprehensive markers in the four species.
Results
Overall dataset
Overall, out of a total of 10,976 potential reactions, 6987 (64%) resulted in ‘any amplification’, and 5711 (52%) produced amplicons within the expected size. ‘Any amplification’ reactions include ones that did not produce any amplicons within 50 bp of the expected range but did produce amplicons outside the range and/or greater than 600 bp. Therefore, 1276 reactions (6987 any amplification—5711 expected size amplifications) only produced amplicons from non-target loci. Twelve percent of the reactions (1361) produced any amplicons outside of the expected bp range. Eight percent or 882 of the reactions produced amplicons greater than 600 bp. The full datasets with allele sizes are broken down by marker set (VD, VF, Vore, and VR; Supplementary Tables 3–6). The maximum number of alleles found per individual was four, despite one species being octaploid. The overall amplification rate for the markers in Viburnum was 60% and 14% in samples outside of the Viburnum genus.
The marker set with the overall highest amplification rate was Vore (Oreinotinus; 66%) and the lowest amplification percentage was VF (V. farreri; 40%). The VD (V. dilatatum; a member of Succotinus subclade and of Laminotinus major clade) markers amplified the most loci in Succotinus (95%), Coriacea (Laminotinus; 83%), and Lobata (Laminotinus; 82%; Table 1). The VF (V. farreri; Solenotinus subclade and Crenotinus major clade) markers amplified the most loci in Solenotinus (70%), Lutescentia (Crenotinus; 68%), and Dentata (Porphyrotinus; 58%). The Vore (Oreinotinus subclade; Porphyrotinus major clade) markers amplified the most loci in Succotinus (Laminotinus; 98%), Dentata (Porphyrotinus; 97%), and Mollotinus (Porphyrotinus; 97%). The VR (V. rufidulum; Lentago subclade and Valvatotinus major clade) markers amplified the most loci in Lentago (94%), Euviburnum (Valvatotinus; 70%), and Dentata (Porphyrotinus; 64%). The frequency of amplification per marker per subclade was also calculated (Fig. 2). The markers developed for the Oreinotinus subclade amplified loci in species the most uniformly across all subclades. To aid in the selection of markers for future studies, overall in range/expected and spurious amplifications were calculated for each marker (Fig. 3). More expected amplifications did not always coincide with fewer spurious amplifications.
Heatmap of frequency of amplification per subclade or family for each of the 49 markers (loci). Vertical grey lines mark the separation of marker set groups and horizontal grey lines mark the separation of major clades and subclades/species not classified into major clades. The darker the square, the higher the amplification frequency of that marker for the sample group.
Overall expected and spurious amplification frequency percentages for all 49 SSR markers. Expected amplification is shown in grey. Spurious amplification is shown in color. Yellow represents the Viburnum dilatatum (VD) markers, green represents the V. farreri (VF) markers, teal represents the Oreinotinus subclade (Vore) markers, and blue represents the V. rufidulum (VR) markers.
Validation: comprehensive SSR marker set
Fourteen markers were selected as a starting point for future research with any Viburnum species, regardless of clade. These markers included VD003, VD004, VD014, VD016, VF20_01, VF20_02, VF20_22, Vore02, Vore04, Vore07, Vore14, Vore15, VR004, and VR011, which displayed high amplification rates across most subclades (Fig. 4). This set of markers successfully amplified loci in 74% of all samples, including 85% of Viburnum species samples and 19% of species not classified in the Viburnum genus. This comprehensive marker set had high amplification frequencies and detected heterozygosity in the four species selected for validation (Table 2). The observed heterozygosity ranged from 0 to 1 with an average of 0.31 in V. carlesii, 0.48 in V. opulus, 0.35 in V. plicatum, and 0.50 in V. tinus. The amplification rates and observed heterozygosity were not as high in the species outside of Viburnum, from which amplification success was an average of 19% in the rest of the Viburnaceae and 20% in Caprifoliaceae. The average observed heterozygosity was 0.10 for the species of the Viburnaceae and 0.01 for species in the Caprifoliaceae.
Discussion
Amplification within expected versus outside size range
The transferability of SSR markers between individuals, species, and genera is dependent upon the conservation of the primer sites. Despite primer sequences being around 20 bp in length, they can bind to locations in the genome other than the target locus, causing spurious banding. This is especially prevalent in cross-amplification studies. Anything greater than 50 bp from the published expected range was considered a different locus than the target one, which happened in markers across all four sets and was not concentrated in a set from a single source. This 50 bp cutoff was selected because the majority of strong peaks with little to no spurious banding occurred within 50 bp of the expected size range. Consequently, some of the amplicons outside of the 50 bp cut-off could be from the target loci. Sequencing the amplicons would resolve this uncertainty, but sequencing was outside the scope of this unusually large study. However, sequencing amplicons is highly recommended in cross-amplification studies due to the potential of homoplasy33,39.
Future research with these markers should involve sequencing amplicons from species of distantly related Viburnum subclades from the marker source species to confirm the expected locus was amplified. Additionally, amplicons that were greater than 50 bp outside of the expected range should be sequenced to confirm they do not contain the expected SSR. This could be especially helpful for potentially using these markers in the Caprifoliaceae and the other species in the Viburnaceae. Many markers produced clean, strong bands in samples from outside of Viburnum, but most amplicons were too far removed from the expected size range to assert they were the correct locus without sequencing. After sequencing, more of the markers could potentially be used for a larger variety of species. Additionally, increasing the annealing temperature in the PCR protocol could help eliminate amplifying non-target loci.
Overall amplification
The transferability of SSR markers throughout plant species overall is lower than in animals43. The cross-amplification success of markers in eudicots is about 71%, with amplification success within genera almost 80% and among genera almost 60%43. A study with nine Rosaceae species determined Prunus SSR markers had cross-species amplifications of 84% within the genus and 38% outside Prunus41. Despite the comparable size of the Prunus genus, with about 200 species48, Viburnum markers displayed much lower cross-amplification with 60% within genus and 14% outside of the genus. However, more than five times the species were included in our study.
The overall trends of marker amplification success aligned with the subclades the markers were developed from. VD markers amplified the highest percentage of target loci in the Laminotinus and Porphyrotinus major clades. VF markers amplified the most in the Crenotinus and Porphyrotinus major clades. VR markers amplified the most in the Valvatotinus and Crenotinus major clades. Additionally, VR markers amplified the highest percentage of target loci in the Viburnaceae and Caprifoliaceae out of the four marker sets. Vore markers had high amplification success in most clades, which is likely due to the SSRs initially being identified in four different species and the markers subsequently developed in two species30. This process was effectively selecting for markers that displayed cross-amplification from the beginning. Despite this, the amplification frequency of Vore markers in the Oreinotinus subclade was low compared to most other subclades. The leaf samples for the Oreinotinus subclade were from herbaria and collected between 1935 and 1998, therefore the sample age is likely a contributing factor to DNA degradation and the resulting low amplification success. The older sample age also likely explains the relatively low amplification success of all 49 makers in the Oreinotinus subclade.
The Vore markers that had particularly low amplification frequency in Oreinotinus were not initially discovered in species in the Oreinotinus subclade30. Vore05 was originally from V. trilobum (Opulus subclade) and had 0% amplification in the Oreinotinus subclade. The Vore16 locus was isolated from V. dentatum (Dentata subclade) and only had 17% amplification in the Oreinotinus subclade. It should be noted Vore09 was mined from mitochondrial NGS data of V. dentatum and despite the marker amplifying loci well and being polymorphic, all samples were inherently homozygous at this locus and special consideration should be taken before use30.
Phylogenetic and morphological classification comparisons
The phylogenetic position of lone species (i.e., V. clemensiae and V. amplificatum) and subclades (i.e., Pseudotinus, and Urceolata) have been changed as additional data has accrued. Viburnum clemensiae has been placed as sister to the rest of Viburnum20,22,24, but recently has been considered sister to a clade containing Crenotinus, Valvatotinus, and subclades Pseudotinus and Urceolata1. VF and VD markers had the highest amplification rate (50% and 45%, respectively) in V. clemensiae. Additionally, V. farreri and V. clemensiae are two of the few species that have panicle-like inflorescence. This could indicate that V. clemensiae would be better suited as sister to the clade containing Crenotinus, Valvatotinus, and subclades Pseudotinus and Urceolata as suggested in Landis et al.1, rather than as sister to the whole genus.
Pseudotinus has been classified as a polytomy with Valvatotinus and rest of Viburnum (Pluriviburnum)22, sister to Valvatotinus20, and sister to Urceolata1,24. Considering the morphological features of buds, leaf margin, inflorescence architecture, and extrafloral nectaries, this subclade shares the most features with V. rufidulum in Valvatotinus22. However, VR markers displayed the lowest amount of amplification to this subclade. The results of this study therefore do not support the placement of Pseudotinus as sister to Valvatotinus, and perhaps sister to Urceolata is more appropriate.
Urceolata has been classified as sister to Amplicrenotinus (Crenotinus + V. amplificatum)20,22 and sister to Pseudotinus1,24. VF (Crenotinus) markers exhibited the lowest amplification success in Urceolata, which supports not classifying Urceolata as sister to Amplicrenotinus. Urceolata does not share many morphological features with any of the species markers were developed from, but interestingly it shares the least morphological features with V. dilatatum22. The only feature they share is the umbel-like inflorescence, which is present in most Viburnum species. Despite this, VD markers had the highest amplification percentage at 39% across Urceolata species.
The taxonomic placement of the Viburnum genus is also under debate and whether the family should be called either Adoxaceae or Viburnaceae, or if they should be separate families3. The amplification frequencies of the markers in the outgroup species of the Viburnaceae and Caprifoliaceae were low and similar, which perhaps suggests the Viburnaceae and Adoxaceae should be separate families, with Sambucus and Adoxa in Adoxaceae.
Validation: comprehensive SSR marker set
The primary goal of this study was to evaluate the cross-amplification success of the 49 developed markers to guide future research and classification in Viburnum. Depending on the application, some markers will have higher amplification rates and polymorphisms than others, but as an initial go-to set, the following 14 markers are recommended: VD003, VD004, VD014, VD016, VF20_01, VF20_02, VF20_22, Vore02, Vore04, Vore07, Vore14, Vore15, VR004, and VR011. If investigating a species outside of Viburnum, markers not included in this comprehensive set would likely have higher amplification success. Adding markers with higher amplification success outside of Viburnum such as VF20_17, VF20_21, Vore10, Vore12, VR028, and VR043 to markers VD014, VD016, VF20_02, VF20_22, Vore04, Vore07, Vore15, and VR011 within the comprehensive set would likely yield better results for studies concerning species not classified in the Viburnum genus.
This comprehensive marker set provides coverage of all subclades and the proof-of-concept species. Similar to the overall study, this subset of the dataset in general also demonstrates that the conservation of the primer site is correlated with the evolutionary distance between the species used in marker development and the sample species. Viburnum carlesii is a member of the Euviburnum subclade and Valvatotinus major clade, which helps to explain VD004 (from Laminotinus major clade) having the lowest amplification rate. Viburnum opulus is a member of the Opulus subclade, which is not classified in any of the four major clades but is more closely related to the Laminotinus major clade, which possibly explains VR004 having the lowest amplification rate.
Despite these comprehensive SSR markers displaying wide cross-amplification, they still also have relatively high observed heterozygosity rates and thus, are useful for genetic diversity studies of any specific species. The high observed heterozygosity rates demonstrate the markers’ ability to identify different alleles in individuals. To the best of our knowledge, this makes this comprehensive marker set the first marker set capable of characterizing any species in a large genus. This will save time and resources in future research as no additional markers will need to be developed. Once sequencing is performed on select amplicons, these comprehensive SSR markers will be ready for use within Viburnum, likely with limited PCR optimization. Future applications for these markers could include characterization and population-level studies for any species within or closely related to Viburnum as well as adding them to established linkage maps49 for future breeding efforts.
Data availability
All data generated or analyzed in this study are included in this published article and its supplementary information files.
References
Landis, M. J. et al. Joint phylogenetic estimation of geographic movements and biome shifts during the global diversification of Viburnum. Syst. Biol. https://doi.org/10.1093/sysbio/syaa027 (2021).
Ulloa Ulloa, C., Acevedo-Rodríguez, P., Beck, S.G.; Belgrano, M.J., Bernal González, R., Berry, P.E., Brako, L., Celis, M., Davidse, G., Gradstein, S.R., et al. An integrated assessment of vascular plants species of the Americas (Online Updates). Available online: www.tropicos.org/Project/VPA (accessed on 19 July 2022).
Applequist, W. L. Report of the nomenclature committee for vascular plants: 65. Taxon 62, 1315–1326. https://doi.org/10.12705/626.49 (2013).
Egolf, D. R. A cytological study of the genus Viburnum. J. Arnold Arboretum 43, 132–172 (1962).
Zhang, N.-N., Sun, W.-B. & Yang, J. Chromosome counts and karyotype analysis of Viburnum taxa (Adoxaceae). Caryologia 69, 12–19. https://doi.org/10.1080/00087114.2015.1109929 (2016).
Winkworth, R. C. & Donoghue, M. J. Viburnum phylogeny: Evidence from the duplicated nuclear gene GBSSI. Mol. Phylogenet. Evol. 33, 109–126. https://doi.org/10.1016/j.ympev.2004.05.006 (2004).
Bai, C., Alverson, W. S., Follansbee, A. & Waller, D. M. New reports of nuclear DNA content for 407 vascular plant taxa from the United States. Ann. Bot. 110, 1623–1629 (2012).
Zhang, L., Cao, B. & Bai, C. New reports of nuclear DNA content for 66 traditional Chinese medicinal plant taxa in China. Caryologia 66, 375–383. https://doi.org/10.1080/00087114.2013.859443 (2013).
Olszewska, M.J., Osiecka, R. The relationship between 2 C DNA content, systematic position, and the level of nuclear DNA endoreplication during differentiation of root parenchyma in some dicotyledonous shrubs and trees. comparison with Herbaceous species. Biochemie und Physiologie der Pflanzen 179, 641–657 (1984). https://doi.org/10.1016/S0015-3796(84)80021-9
Pustahija, F. et al. Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa. Plant Soil 373, 427–453 (2013).
Moeglein, M. K., Chatelet, D. S., Donoghue, M. J. & Edwards, E. J. Evolutionary dynamics of genome size in a radiation of woody plants. Am. J. Bot. 107, 1527–1541. https://doi.org/10.1002/ajb2.1544 (2020).
Fridley, J. D. & Craddock, A. Contrasting growth phenology of native and invasive forest shrubs mediated by genome size. New Phytol. 207, 659–668. https://doi.org/10.1111/nph.13384 (2015).
Cao, B., Zhang, L.-L. & Bai, C.-K. Estimation of nuclear DNA content in tannin-rich medicinal plant Cornus officinalis by flow cytometry. Chin. Herb. Med. 6, 152–158. https://doi.org/10.1016/S1674-6384(14)60024-1 (2014).
Zonneveld, B., Leitch, I. & Bennett, M. First nuclear DNA amounts in more than 300 angiosperms. Ann. Bot. 96, 229–244. https://doi.org/10.1093/aob/mci170 (2005).
Jedrzejczyk, I. & Sliwinska, E. Leaves and seeds as materials for flow cytometric estimation of the genome size of 11 Rosaceae woody species containing DNA-staining inhibitors. J. Bot. (2010). https://doi.org/10.1155/2010/930895.
Dickson, E., Arumuganathan, K., Kresovich, S. & Doyle, J. Nuclear DNA content variation within the Rosaceae. Am. J. Bot. 79, 1081–1086. https://doi.org/10.2307/2444917 (1992).
Dirr, M. Viburnums: Flowering Shrubs for Every Season (Timber Press, 2007).
USDA-NASS. 2019 Census of horticultural specialities: Table 23. Deciduous shrubs (2020).
Spriggs, E. L. et al. Differences in flowering time maintain species boundaries in a continental radiation of Viburnum. Am. J. Bot. 106, 833–849. https://doi.org/10.1002/ajb2.1292 (2019).
Spriggs, E. L. et al. Temperate radiations and dying embers of a tropical past: The diversification of Viburnum. New Phytol. 207, 340–354. https://doi.org/10.1111/nph.13305 (2015).
Winkworth, R. C. & Donoghue, M. J. Viburnum phylogeny based on combined molecular data: Implications for taxonomy and biogeography. Am. J. Bot. 92, 653–666. https://doi.org/10.3732/ajb.92.4.653 (2005).
Clement, W. L., Arakaki, M., Sweeney, P. W., Edwards, E. J. & Donoghue, M. J. A chloroplast tree for Viburnum (Adoxaceae) and its implications for phylogenetic classification and character evolution. Am. J. Bot. 101, 1029–1049. https://doi.org/10.3732/ajb.1400015 (2014).
Choi, Y. G., Youm, J. W., Lim, C. E. & Oh, S.-H. Phylogenetic analysis of Viburnum (Adoxaceae) in Korea using DNA sequences. Korean J. Pl. Taxon 48, 206–217. https://doi.org/10.11110/kjpt.2018.48.3.206 (2018).
Eaton, D. A. R., Spriggs, E. L., Park, B. & Donoghue, M. J. Misconceptions on missing data in RAD-seq phylogenetics with a deep-scale example from flowering plants. Syst. Biol. 66, 399–412. https://doi.org/10.1093/sysbio/syw092 (2017).
Clement, W. L. & Donoghue, M. J. Barcoding success as a function of phylogenetic relatedness in Viburnum, a clade of woody angiosperms. BMC Evol. Biol. 12, 73. https://doi.org/10.1186/1471-2148-12-73 (2012).
Donoghue, M. J., Baldwin, B. G., Li, J. & Winkworth, R. C. Viburnum phylogeny based on chloroplast trnK intron and nuclear ribosomal ITS DNA sequences. Syst. Bot. 29(188–198), 111 (2004).
Spriggs, E. L. et al. Restriction-site-associated DNA sequencing reveals a cryptic Viburnum species on the North American Coastal Plain. Syst. Biol. 68, 187–203. https://doi.org/10.1093/sysbio/syy084 (2019).
Dean, D. et al. Screening and characterization of 11 novel microsatellite markers from Viburnum dilatatum. HortScience 46, 1456–1459. https://doi.org/10.21273/hortsci.46.11.1456 (2011).
Dean, D. et al. Analysis of genetic diversity and population structure for the native tree Viburnum rufidulum occurring in Kentucky and Tennessee. J. Am. Soc. Hortic. Sci. 140, 523. https://doi.org/10.21273/jashs.140.6.523 (2015).
Barish, S., Arakaki, M., Edwards, E. J., Donoghue, M. J. & Clement, W. L. Characterization of 16 microsatellite markers for the Oreinotinus clade of Viburnum (Adoxaceae). Appl. Plant Sci. 4, 1600103. https://doi.org/10.3732/apps.1600103 (2016).
Hamm, T. P. et al. Development and characterization of 15 novel genomic SSRs for Viburnum farreri. Plants 10, 487. https://doi.org/10.3390/plants10030487 (2021).
Powell, W., Machray, G. C. & Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1, 215–222. https://doi.org/10.1016/1360-1385(96)86898-1 (1996).
Peakall, R., Gilmore, S., Keys, W., Morgante, M. & Rafalski, A. Cross-species amplification of soybean (Glycine max) simple sequence repeats (SSRs) within the genus and other legume genera: Implications for the transferability of SSRs in plants. Mol. Biol. Evol. 15, 1275–1287. https://doi.org/10.1093/oxfordjournals.molbev.a025856 (1998).
Gupta, P. K. & Varshney, R. K. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113, 163–185. https://doi.org/10.1023/A:1003910819967 (2000).
Ony, M. A. et al. Habitat fragmentation influences genetic diversity and differentiation: Fine-scale population structure of Cercis canadensis (eastern redbud). Ecol. Evol. 10, 3655–3670. https://doi.org/10.1002/ece3.6141 (2020).
Manechini, J. R. V., da Costa, J. B., Pereira, B. T., Carlini-Garcia, L. A., Xavier, M. A., Landell, M.G.d.A., Pinto, L. R. Unraveling the genetic structure of Brazilian commercial sugarcane cultivars through microsatellite markers. PLoS ONE 13, 0195623 (2018). https://doi.org/10.1371/journal.pone.0195623.
Scariot, V., Akkak, A. & Botta, R. Characterization and genetic relationships of wild species and old garden roses based on microsatellite analysis. J. Am. Soc. Hortic. Sci. 131, 66. https://doi.org/10.21273/jashs.131.1.66 (2006).
Stackhouse, T. et al. Conventional gel electrophoresis and TaqMan probes enable rapid confirmation of thousand cankers disease from diagnostic samples. Plant Dis. 105, 3171–3180. https://doi.org/10.1094/pdis-10-20-2258-re (2021).
Wadl, P. A., Wang, X., Moulton, J. K., Hokanson, S. C., Skinner, J. A., Rinehart, T. A., Reed, S. M., Pantalone, V. R., Trigiano, R. N. Transfer of Cornus florida and C. kousa simple sequence repeats to selected Cornus (Cornaceae) species. J. Am. Soc. Hortic. Sci. 135, 279 (2010). https://doi.org/10.21273/jashs.135.3.279.
Wadl, P. A., Trigiano, R. N., Werner, D. J., Pooler, M. R. & Rinehart, T. A. Simple sequence repeat markers from Cercis canadensis show wide cross-species transfer and use in genetic studies. J. Am. Soc. Hortic. Sci. 137, 189. https://doi.org/10.21273/jashs.137.3.189 (2012).
Mnejja, M., Garcia-Mas, J., Audergon, J.-M. & Arús, P. Prunus microsatellite marker transferability across rosaceous crops. Tree Genet. Genomes 6, 689–700. https://doi.org/10.1007/s11295-010-0284-z (2010).
Aiello, D. et al. Evaluation of cross-species transferability of SSR markers in Foeniculum vulgare. Plants 9, 175. https://doi.org/10.3390/plants9020175 (2020).
Barbara, T. et al. Cross-species transfer of nuclear microsatellite markers: Potential and limitations. Mol. Ecol. 16, 3759–3767. https://doi.org/10.1111/j.1365-294X.2007.03439.x (2007).
Dean, D. Assessing the genetic diversity of the genus Viburnum using simple sequence repeats (University of Tennessee, 2014).
Paulauskas, A. et al. Differentiation of Viburnum accessions according to their molecular, biochemical, genotoxic and microbiological features of importance to selection. Acad. J. Agric. Res. 3, 081–093. https://doi.org/10.15413/ajar.2015.0121 (2015).
Diversity Arrays Technology. Plant DNA extraction protocol for DArT. Available online: https://www.diversityarrays.com/orderinstructions/plant-dna-extraction-protocol-for-dart/ (accessed on 22 July 2022).
Don, R. H., Cox, P. T., Wainwright, B. J., Baker, K. & Mattick, J. S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19, 4008–4008. https://doi.org/10.1093/nar/19.14.4008 (1991).
Potter, D. Prunus. In Wild Crop Relatives: Genomic and Breeding Resources: Temperate Fruits, Kole, C., Ed.; Springer Berlin Heidelberg: Berlin, Heidelberg, 2011; pp. 129–145.
Al-Niemi, T., Weeden, N. F., McCown, B. H. & Hoch, W. A. Genetic analysis of an interspecific cross in ornamental viburnum (Viburnum). J. Hered. 103, 2–12. https://doi.org/10.1093/jhered/esr133 (2011).
Acknowledgements
The authors gratefully recognize the U.S. National Arboretum Herbarium, The Morton Arboretum, U.S. National Arboretum, Yale Peabody Museum, Herbarium of the Arnold Arboretum, Arnold Arboretum, Spring Grove Cemetery and Arboretum, Mt. Airy Arboretum, JC Raulston Arboretum, Morris Arboretum, University of Tennessee Herbarium, Carnegie Museum of Natural History Herbarium, University of Washington Botanical Garden, and A.C. Moore Herbarium for the donation of plant material for this study. We also thank Shade Niece for his technical help. This work was supported by a Non-Assistance Cooperative Agreement between the University of Tennessee and USDA, ARS (NACA 58-6062-6).
Author information
Authors and Affiliations
Contributions
Conceptualization, R.N.T. and T.P.H.; methodology, R.N.T., T.P.H. and S.L.B.; software, T.P.H.; validation, T.P.H. and S.L.B.; formal analysis, T.P.H.; investigation, T.P.H. and M.N.; resources, T.P.H., R.N.T., M.N., T.G.R. and S.L.B.; data curation, T.P.H., S.L.B.; writing—original draft preparation, T.P.H.; writing—review and editing, R.N.T., M.N., S.L.B., T.G.R.; visualization, T.P.H.; supervision, T.P.H. and S.L.B.; project administration, T.P.H.; funding acquisition, R.N.T. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hamm, T.P., Nowicki, M., Boggess, S.L. et al. A set of SSR markers to characterize genetic diversity in all Viburnum species. Sci Rep 13, 5343 (2023). https://doi.org/10.1038/s41598-023-31878-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-31878-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.