Abstract
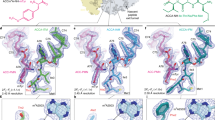
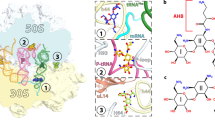
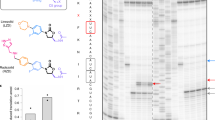
Viomycin and capreomycin belong to the tuberactinomycin family of antibiotics, which are among the most effective antibiotics against multidrug-resistant tuberculosis. Here we present two crystal structures of the 70S ribosome in complex with three tRNAs and bound to either viomycin or capreomycin at 3.3- and 3.5-Å resolution, respectively. Both antibiotics bind to the same site on the ribosome, which lies at the interface between helix 44 of the small ribosomal subunit and helix 69 of the large ribosomal subunit. The structures of these complexes suggest that the tuberactinomycins inhibit translocation by stabilizing the tRNA in the A site in the pretranslocation state. In addition, these structures show that the tuberactinomycins bind adjacent to the binding sites for the paromomycin and hygromycin B antibiotics, which may enable the development of new derivatives of tuberactinomycins that are effective against drug-resistant strains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Guy, E.S. & Mallampalli, A. Managing TB in the 21st century: existing and novel drug therapies. Ther Adv Respir Dis 2, 401–408 (2008).
Hugonnet, J.E., Tremblay, L.W., Boshoff, H.I., Barry, C.E. III & Blanchard, J.S. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 323, 1215–1218 (2009).
Makarov, V. et al. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324, 801–804 (2009).
WHO. Global tuberculosis control: epidemology, strategy, financing (WHO report 2009) 314 (World Health Organization, Geneva, Switzerland, 2009).
Yamada, T., Mizugichi, Y., Nierhaus, K.H. & Wittmann, H.G. Resistance to viomycin conferred by RNA of either ribosomal subunit. Nature 275, 460–461 (1978).
Thomas, M.G., Chan, Y.A. & Ozanick, S.G. Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob. Agents Chemother. 47, 2823–2830 (2003).
Johansen, S.K., Maus, C.E., Plikaytis, B.B. & Douthwaite, S. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell 23, 173–182 (2006).
Herr, E.B. Jr. & Redstone, M.O. Chemical and physical characterization of capreomycin. Ann. NY Acad. Sci. 135, 940–946 (1966).
Modolell, J. & Vazquez, D. The inhibition of ribosomal translocation by viomycin. Eur. J. Biochem. 81, 491–497 (1977).
Ogle, J.M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001).
Schmeing, T.M., Huang, K.S., Strobel, S.A. & Steitz, T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438, 520–524 (2005).
Voorhees, R.M., Weixlbaumer, A., Loakes, D., Kelley, A.C. & Ramakrishnan, V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 16, 528–533 (2009).
Lancaster, L.E., Savelsbergh, A., Kleanthous, C., Wintermeyer, W. & Rodnina, M.V. Colicin E3 cleavage of 16S rRNA impairs decoding and accelerates tRNA translocation on Escherichia coli ribosomes. Mol. Microbiol. 69, 390–401 (2008).
Nomoto, S. & Shiba, T. Chemical studies on tuberactinomycin. XIII. Modification of β-ureidodehydroalanine residue in tuberactinomycin N. J. Antibiot. (Tokyo) 30, 1008–1011 (1977).
Yamada, T., Teshima, T. & Shiba, T. Activity of di-β-lysyl-capreomycin IIA and palmitoyl tuberactinamine N against drug-resistant mutants with altered ribosomes. Antimicrob. Agents Chemother. 20, 834–836 (1981).
von Nussbaum, F., Brands, M., Hinzen, B., Weigand, S. & Habich, D. Antibacterial natural products in medicinal chemistry—exodus or revival? Angew. Chem. 45, 5072–5129 (2006).
Monshupanee, T., Gregory, S.T., Douthwaite, S., Chungjatupornchai, W. & Dahlberg, A.E. Mutations in conserved helix 69 of 23S rRNA of Thermus thermophilus that affect capreomycin resistance but not posttranscriptional modifications. J. Bacteriol. 190, 7754–7761 (2008).
Schuwirth, B.S. et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834 (2005).
Ali, I.K., Lancaster, L., Feinberg, J., Joseph, S. & Noller, H.F. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell 23, 865–874 (2006).
Kipper, K., Hetenyi, C., Sild, S., Remme, J. & Liiv, A. Ribosomal intersubunit bridge B2a is involved in factor-dependent translation initiation and translational processivity. J. Mol. Biol. 385, 405–422 (2009).
O'Connor, M. Helix 69 in 23S rRNA modulates decoding by wild type and suppressor tRNAs. Mol. Genet. Genomics 282, 372–380 (2009).
Borovinskaya, M.A. et al. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 14, 727–732 (2007).
Mears, J.A. et al. Modeling a minimal ribosome based on comparative sequence analysis. J. Mol. Biol. 321, 215–234 (2002).
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
Weixlbaumer, A. et al. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat. Struct. Mol. Biol. 14, 733–737 (2007).
Laurberg, M. et al. Structural basis for translation termination on the 70S ribosome. Nature 454, 852–857 (2008).
Korostelev, A. et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. USA 105, 19684–19689 (2008).
Weixlbaumer, A. et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322, 953–956 (2008).
Moazed, D. & Noller, H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148 (1989).
Agirrezabala, X. et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol. Cell 32, 190–197 (2008).
Kim, H.D., Puglisi, J.D. & Chu, S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys. J. 93, 3575–3582 (2007).
Pan, D., Kirillov, S.V. & Cooperman, B.S. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell 25, 519–529 (2007).
Ermolenko, D.N. et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 14, 493–497 (2007).
Cornish, P.V., Ermolenko, D.N., Noller, H.F. & Ha, T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell 30, 578–588 (2008).
Zhang, W., Dunkle, J.A. & Cate, J.H. Structures of the ribosome in intermediate states of ratcheting. Science 325, 1014–1017 (2009).
Shoji, S., Walker, S.E. & Fredrick, K. Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem. Biol. 4, 93–107 (2009).
Peske, F., Savelsbergh, A., Katunin, V.I., Rodnina, M.V. & Wintermeyer, W. Conformational changes of the small ribosomal subunit during elongation factor G–dependent tRNA-mRNA translocation. J. Mol. Biol. 343, 1183–1194 (2004).
Carter, A.P. et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407, 340–348 (2000).
Borovinskaya, M.A., Shoji, S., Fredrick, K. & Cate, J.H. Structural basis for hygromycin B inhibition of protein biosynthesis. RNA 14, 1590–1599 (2008).
Szaflarski, W. et al. New features of the ribosome and ribosomal inhibitors: non-enzymatic recycling, misreading and back-translocation. J. Mol. Biol. 380, 193–205 (2008).
Mayer, C. & RajBhandary, U.L. Conformational change of Escherichia coli initiator methionyl-tRNA(fMet) upon binding to methionyl-tRNA formyl transferase. Nucleic Acids Res. 30, 2844–2850 (2002).
Blaha, G., Stanley, R.E. & Steitz, T.A. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science 325, 966–970 (2009).
Perona, J.J., Swanson, R., Steitz, T.A. & Soll, D. Overproduction and purification of Escherichia coli tRNA(2Gln) and its use in crystallization of the glutaminyl-tRNA synthetase-tRNA(Gln) complex. J. Mol. Biol. 202, 121–126 (1988).
Kabsch, W. XDS. in International Tables in Crystallography, vol. F (eds. Rossmann, M.G. & Arnold, E.) 730–734 (published for the International Union of Crystallography by Springer, Dordrecht, The Netherlands, 2005).
McCoy, A.J., Grosse-Kunstleve, R.W., Adams, P.D., Winn, M.D., Storoni, L.C. & Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Brunger, A.T. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 (2007).
Brunger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Bycroft, B.W. Crystal structure of viomycin, a tuberculostatic antibiotic. JCS Chem. Commun. 660–661 (1972).
Yoshioka, H. et al. Chemical studies on tuberactinomycin. II. Structure of tuberactinomycin O. Tetrahedr. Lett. 2043–2046 (1971).
Bowers, K.J. et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the ACM/IEEE Conference on Supercomputing (ACM/IEEE, 2006).
Jorgensen, W.L., Maxwell, D.S. & Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 (1996).
Jorgensen, W.L., Chandrasekhar, J., Madura, J.D., Impey, R.W. & Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
DeLano, W.L. The PyMol Molecular Graphics System. (DeLano Scientific, Palo Alto, California, USA, 2002).
Acknowledgements
We thank the staffs at the Advanced Photon Source beamline 24-ID and at the National Synchrotron Light Source beamline X29 for help during data collection, the staff at the Center for Structural Biology at Yale University for computational support, I. Lomakin (Yale University) for providing us with methionyl-tRNA synthetase and U.L. RajBhandary (Massachusetts Institute of Technology), K.H. Nierhaus (Max-Planck-Institut für Molekulare Genetik), P. Nissen (Aarhaus University) and M. Sprinzl (University Bayreuth) for providing us with ET-Ts for the overexpression plasmids of methionyl-tRNA formyltransferase, initiator tRNA, EF-Tu and EF-Ts, respectively. This work was supported by the US National Institutes of Health grant GM 22778 to T.A.S.
Author information
Authors and Affiliations
Contributions
R.E.S. prepared and crystallized the complexes; R.E.S. and G.B. collected data and processed and refined X-ray data; G.B. and R.L.G. purified all components of complex; M.D.S. energy-minimized the initial tuberactinomycin structures and solved all computational problems related to the large size of the datasets and complexes; T.A.S., G.B. and R.E.S. contributed to the experimental design and analysis of the structure; T.A.S., G.B. and R.E.S. wrote the manuscript, on which all authors commented.
Corresponding author
Ethics declarations
Competing interests
T.A.S. owns stock in and is on the advisory board of Rib-X Pharmaceuticals, Inc., which does structure-based drug design targeted at the ribosome.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 (PDF 183 kb)
Rights and permissions
About this article
Cite this article
Stanley, R., Blaha, G., Grodzicki, R. et al. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol 17, 289–293 (2010). https://doi.org/10.1038/nsmb.1755
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1755
This article is cited by
-
Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin
Communications Biology (2023)
-
Accuracy mechanism of eukaryotic ribosome translocation
Nature (2021)
-
Translation error clusters induced by aminoglycoside antibiotics
Nature Communications (2021)
-
Mechanisms of drug interactions between translation-inhibiting antibiotics
Nature Communications (2020)
-
New antituberculous drugs derived from natural products: current perspectives and issues in antituberculous drug development
The Journal of Antibiotics (2018)