Key Points
-
Myelodysplastic syndromes (MDS) are a group of diseases that present with the paradox of cytopenia despite a cellular bone marrow. This can be explained by excessive cytokine-mediated intramedullary apoptosis of haematopoietic cells.
-
Emerging biological insights are providing a molecular basis for the phenotypic and clinical heterogeneity of MDS.
-
Defects in ribosomal biogenesis have been associated with both congenital and acquired anaemias. These defects lead to a ribosomal stress response in the cell with an increase in p53 expression, thus providing one molecular explanation for the excessive apoptosis of MDS cells.
-
Abnormal regulation of haematopoiesis by microRNAs (miRNAs) has now been shown to contribute to the pathology of MDS.
-
The importance of epigenetic changes in MDS — initially suspected because of the responsiveness of patients with MDS to demethylating agents — has been molecularly demonstrated through the identification of mutations in genes controlling chromatin methylation and deacetylation.
-
Multiple oncogenes and tumour suppressor genes are mutated, although none has been specifically associated with MDS.
-
Mutations of specific spliceosome genes, either alone or combined with other genetic abnormalities, may be responsible for different MDS phenotypes.
-
An incremental understanding of molecular lesions accounting for clonal expansion in the presence of excessive apoptosis — which is the paradox in MDS — will be the roadmap to improved and targeted therapies.
Abstract
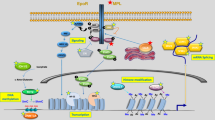
Myelodysplastic syndromes (MDS) are malignant clonal disorders of haematopoietic stem cells and their microenvironment, affecting older individuals (median age ∼70 years). Unique features that are associated with MDS — but which are not necessarily present in every patient with MDS — include excessive apoptosis in maturing clonal cells, a pro-inflammatory bone marrow microenvironment, specific chromosomal abnormalities, abnormal ribosomal protein biogenesis, the presence of uniparental disomy, and mutations affecting genes involved in proliferation, methylation and epigenetic modifications. Although emerging insights establish an association between molecular abnormalities and the phenotypic heterogeneity of MDS, their origin and progression remain enigmatic.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Anastasi, J. et al. Cytogenetic clonality in myelodysplastic syndromes studied with fluorescence in situ hybridization: lineage, response to growth factor therapy, and clone expansion. Blood 81, 1580–1585 (1993).
Raza, A. et al. Apoptosis in bone marrow biopsy samples involving stromal and hematopoietic cells in 50 patients with myelodysplastic syndromes. Blood 86, 268–276 (1995). The first study to report the presence of increased apoptosis in MDS bone marrows as a unique feature of the disease.
Raza, A. et al. Novel insights into the biology of myelodysplastic syndromes: excessive apoptosis and the role of cytokines. Int. J. Hematol. 63, 265–278 (1996).
Parker, J. E. et al. 'Low-risk' myelodysplastic syndrome is associated with excessive apoptosis and an increased ratio of pro- versus anti-apoptotic bcl-2-related proteins. Br. J. Haematol. 103, 1075–1082 (1998).
Parker, J. E. et al. The role of apoptosis, proliferation, and the Bcl-2-related proteins in the myelodysplastic syndromes and acute myeloid leukemia secondary to MDS. Blood 96, 3932–3938 (2000).
Bianco, P. Bone and the hematopoietic niche: a tale of two stem cells. Blood 117, 5281–5288 (2011).
Raaijmakers, M. H. et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature 464, 852–857 (2010). This paper reports that the disruption of Dicer1 or Sbds in osteoprogenitor cells results in myelodysplasia and leukaemia in a mouse model. It is the first study to show that stromal cell dysfunction can result in an MDS-like phenotype.
Raza, A. et al. Cell cycle kinetic studies in 68 patients with myelodysplastic syndromes following intravenous iodo- and/or bromodeoxyuridine. Exp. Hematol. 25, 530–535 (1997).
Mundle, S. D. et al. Indication of an involvement of interleukin-1ß converting enzyme (ICE)-like protease in intramedullary apoptotic cell death in the bone marrows of patients with myelodysplastic syndromes (MDS). Blood 88, 2640–2647 (1996).
Raza, A., et al. A paradigm shift in myelodysplastic syndromes. Leukemia 10, 1648–1652 (1996).
Calado, R. T. Immunologic aspects of hypoplastic myelodysplastic syndrome. Semin. Oncol. 38, 667–672 (2011).
Marisavljevic, D. et al. Hypocellular myelodysplastic syndromes: clinical and biological significance. Med. Oncol. 22, 169–175 (2005).
Wlodarski, M. W. et al. Molecular strategies for detection and quantitation of clonal cytotoxic T-cell responses in aplastic anemia and myelodysplastic syndrome. Blood 108, 2632–2641 (2006).
Sloand, E. M. et al. CD34 cells from patients with trisomy 8 myelodysplastic syndrome (MDS) express early apoptotic markers but avoid programmed cell death by up-regulation of antiapoptotic proteins. Blood 109, 2399–2405 (2007).
Marsh, J. et al. Prospective randomized multicenter study comparing cyclosporin alone versus the combination of antithymocyte globulin and cyclosporin for treatment of patients with nonsevere aplastic anemia: a report from the European Blood and Marrow Transplant (EBMT) Severe Aplastic Anaemia Working Party. Blood 93, 2191–2195 (1999).
Sloand, E. M. et al. Alemtuzumab treatment of intermediate-1 myelodysplasia patients is associated with sustained improvement in blood counts and cytogenetic remissions. J. Clin. Oncol. 28, 5166–5173 (2010).
Horikawa, K. et al. Apoptosis resistance of blood cells from patients with paroxysmal nocturnal hemoglobinuria, aplastic anemia, and myelodysplastic syndrome. Blood 90, 2716–2722 (1997).
Hengartner, M. O. The biochemistry of apoptosis. Nature 407, 770–776 (2000).
Muñoz-Pinedo, C. Signaling pathways that regulate life and cell death: evolution of apoptosis in the context of self-defense. Adv. Exp. Med. Biol. 738, 124–143 (2012).
Kischkel, F. C. et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14, 5579–5588 (1995).
Scott, F. L. et al. The Fas–FADD death domain complex structure unravels signalling by receptor clustering. Nature 457, 1019–1022 (2009).
Dickens, L. S., Powley, I. R., Hughes, M. A. & Macfarlane, M. The 'complexities' of life and death: death receptor signalling platforms. Exp. Cell Res. 318, 1269–1277 (2012).
Mundle, S. D. et al. Evidence for involvement of tumor necrosis factor-α in apoptotic death of bone marrow cells in myelodysplastic syndromes. Am. J. Hematol. 60, 36–47 (1999).
Claessens, Y. E. et al. In vitro proliferation and differentiation of erythroid progenitors from patients with myelodysplastic syndromes: evidence for Fas-dependent apoptosis. Blood 99, 1594–1601 (2002).
Campioni, D. et al. Evidence for a role of TNF-related apoptosis-inducing ligand (TRAIL) in the anemia of myelodysplastic syndromes. Am. J. Pathol. 166, 557–563 (2005).
Ali, A. et al. Sequential activation of caspase-1 and caspase-3-like proteases during apoptosis in myelodysplastic syndromes. J. Hematother. Stem Cell Res. 8, 343–356 (1999).
Hellström-Lindberg, E. et al. Apoptosis in refractory anaemia with ringed sideroblasts is initiated at the stem cell level and associated with increased activation of caspases. Br. J. Haematol. 112, 714–726 (2001).
Mundle, S. D. et al. Correlation of tumor necrosis factor-α (TNFα) with high Caspase 3-like activity in myelodysplastic syndromes. Cancer Lett. 140, 201–207 (1999).
Shimazaki, K., Ohshima, K., Suzumiya, J., Kawasaki, C. & Kikuchi, M. Evaluation of apoptosis as a prognostic factor in myelodysplastic syndromes. Br. J. Haematol. 110, 584–590 (2000).
Fatica, A. & Tollervey, D. Making ribosomes. Curr. Opin. Cell Biol. 14, 313–318 (2002).
van Riggelen, J., Yetil, A. & Felsher, D. W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nature Rev. Cancer. 10, 301–309 (2010).
Draptchinskaia, N. et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nature Genet. 21, 169–175 (1999).
Heiss, N. S. et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature Genet. 19, 32–38 (1998).
Ridanpää, M. et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 104, 195–203 (2001).
Boocock, G. R. et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nature Genet. 33, 97–101 (2003).
Ebert, B. L., et al. Identification of RPS14 as a 5q−- syndrome gene by RNA interference screen. Nature 451, 335–339 (2008). This study shows that haploinsufficiency for a ribosomal protein gene ( RPS14 ) contributes to the phenotype of 5q− syndrome.
Starczynowski, D. T. et al. Identification of miR-145 and miR-146a as mediators of the 5q− syndrome phenotype. Nature Med. 16, 49–58 (2010). This study reports on the contribution of two miRNAs that are found in the common deleted region of 5q− syndrome to the unique features of this disease.
Kumar, M. S. et al. Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5q− syndrome. Blood 118, 4666–4673 (2011).
Pellagatti, A. et al. Haploinsufficiency of RPS14 in 5q− syndrome is associated with deregulation of ribosomal- and translation-related genes. Br. J. Haematol. 142, 57–64 (2008).
Vousden, K. H. & Prives, C. Blinded by the light: the growing complexity of p53. Cell 137, 413–431 (2009).
Barlow, J. L. et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q− syndrome. Nature Med. 16, 59–66 (2010).
Kruse, J. P. & Gu, W. Modes of p53 regulation. Cell 137, 609–622 (2009).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell. 136, 215–233 (2009).
Khraiwesh, B. et al. Transcriptional control of gene expression by microRNAs. Cell 140, 111–122 (2010).
Calin, G. A. et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA 101, 11755–11760 (2004).
Garzon, R. et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 111, 3183–3189 (2008).
Hussein, K. et al. Aberrant microRNA expression pattern in myelodysplastic bone marrow cells. Leuk. Res. 34, 1169–1174 (2010).
Sokol, L. et al. Identification of a risk dependent microRNA expression signature in myelodysplastic syndromes. Br. J. Haematol. 153, 24–32 (2011).
Erdogan, B. et al. Diagnostic microRNAs in myelodysplastic syndrome. Exp. Hematol. 39, 915–926 (2011).
Dostalova, M. et al. Distinctive microRNA expression profiles in CD34+ bone marrow cells from patients with myelodysplastic syndrome. Eur. J. Hum. Genet. 19, 313–319 (2011).
Votavova, H. et al. Differential expression of microRNAs in CD34+ cells of 5q− syndrome. J. Hematol. Oncol. 4, 1 (2010).
Hussein, K. et al. Expression of myelopoiesis-associated microRNA in bone marrow cells of atypical chronic myeloid leukaemia and chronic myelomonocytic leukaemia. Ann. Hematol. 90, 307–313 (2010).
Pons, A. et al. Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk. Lymphoma 50, 1854–1859 (2009).
Rhyasen, G. W. & Starczynowski, D. T. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia 26, 13–22 (2012).
Boldin, M. P. et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 208, 1189–1201 (2011).
Galili, N. et al. Prediction of response to therapy with ezatiostat in lower risk myelodysplastic syndrome. J. Hematol. Oncol. 5, 20 (2012).
Ley, T. J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010). The first report to show that the methyltransferase gene DNMT3A is a recurrent mutation in de novo AML and is independently associated with poor outcome.
Ewalt, M. et al. DNMT3a mutations in high-risk myelodysplastic syndrome parallel those found in acute myeloid leukemia. Blood Cancer J. 1, e9 (2011).
Walter, M. J. et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 25, 1153–1158 (2011).
Ko, M. et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468, 839–843 (2010). This study demonstrates that TET2 mutations favour the development of myeloid neoplasms.
Moran-Crusio, K. et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20, 11–24 (2011).
Li, Z. et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118, 4509–4518 (2011).
Delhommeau, F. et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 360, 2289–2301 (2009).
Kosmider, O. et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs). Blood 114, 3285–3291 (2009).
Smith, A. E. et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood 116, 3923–3932 (2010).
Kosmider, O. et al. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia. 24, 1094–1096 (2010).
Thol, F. et al. IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica 95, 1668–1674 (2010).
Figueroa, M. E. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010).
Cho, Y. S., Kim, E. J., Park, U. H., Sin, H. S. & Um, S. J. Additional sex comb-like 1 (ASXL1), in cooperation with SRC-1, acts as a ligand-dependent coactivator for retinoic acid receptor. J. Biol. Chem. 281, 17588–17598 (2006).
Lee, S. W. et al. ASXL1 represses retinoic acid receptor-mediated transcription through associating with HP1 and LSD1. J. Biol. Chem. 285, 18–29 (2010).
Fisher, C. L. et al. Loss-of-function Additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood 115, 38–46 (2010).
Boultwood, J. et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia 24, 1062–1065 (2010).
Thol, F. et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J. Clin. Oncol. 29, 2499–2506 (2011).
Nikoloski, G. et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nature Genet. 42, 665–667 (2010).
Ernst, T. et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature Genet. 42, 722–726 (2010).
Rowley, J. D. Identification of a translocation with quinacrine fluorescence in a patient with acute leukemia. Ann. Genet. 16, 109–112 (1973).
Xiao, Z. et al. Molecular characterization of genomic AML1-ETO fusions in childhood leukemia. Leukemia. 15, 1906–1913 (2001).
Harada, H. et al. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood 103, 2316–2324 (2004).
Song, W. J. et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nature Genet. 23, 166–175 (1999).
Watanabe-Okochi, et al. AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood 111, 4297–4308 (2008).
Tang, J. L. et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood 114, 5352–5361 (2009).
Bejar, R. et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 364, 2496–2506 (2011). This study reports the somatic mutations found in MDS and the poor prognostic consequences of TP53, EZH2, ETV6, RUNX1 and ASXL mutations.
Kita-Sasai, Y. et al. International prognostic scoring system and TP53 mutations are independent prognostic indicators for patients with myelodysplastic syndrome. Br. J. Haematol. 115, 309–312 (2001).
Haferlach, C. et al. The inv(3)(q21q26)/t(3;3)(q21;q26) is frequently accompanied by alterations of the RUNX1, KRAS and NRAS and NF1 genes and mediates adverse prognosis both in MDS and in AML: a study in 39 cases of MDS or AML. Leukemia. 25, 874–877 (2011).
Szpurka, H. et al. Refractory anemia with ringed sideroblasts associated with marked thrombocytosis (RARS-T), another myeloproliferative condition characterized by JAK2 V617F mutation. Blood 108, 2173–2181 (2006).
Lea, N. C. et al. The JAK2 V617F mutation is rare in RARS but common in RARS-T. Leukemia 20, 2060–2061 (2006).
Malcovati, L. et al. Molecular and clinical features of refractory anemia with ringed sideroblasts associated with marked thrombocytosis. Blood 114, 3538–3545 (2009).
Zikria, J., Galili, N., Tsai, W. Y. & Raza, A. Thrombocytosis in myelodysplastic syndromes: not an innocent bystander. J. Blood Disord. Transfus. S3:002 (2012).
Bejar, R. et al. Validation of a prognostic model and the impact of mutations in 288 patients with lower risk myelodysplastic syndromes. J. Clin. Oncol. 30, 3376–3382 (2012).
Mohamedali, A. et al. Prevalence and prognostic significance of allelic imbalance by single-nucleotide polymorphism analysis in low-risk myelodysplastic syndromes. Blood 110, 3365–3373 (2007).
Gondek, L. P. et al. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood 111, 1534–1542 (2008).
Heinrichs, S. et al. Accurate detection of uniparental disomy and microdeletions by SNP array analysis in myelodysplastic syndromes with normal cytogenetics. Leukemia 23, 1605–1613 (2009).
Tiu, R. V. et al. Prognostic impact of SNP array karyotyping in myelodysplastic syndromes and related myeloid malignancies. Blood 117, 4552–4560 (2011).
Evers, C. et al. Molecular definition of chromosome arm 5q deletion end points and detection of hidden aberrations in patients with myelodysplastic syndromes and isolated del(5q) using oligonucleotide array CGH. Genes Chromosomes Cancer 46, 1119–1128 (2007).
Starczynowski, D. T. et al. High-resolution whole genome tiling path array CGH analysis of CD34+ cells from patients with low-risk myelodysplastic syndromes reveals cryptic copy number alterations and predicts overall and leukemia-free survival. Blood 112, 3412–3424 (2008).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011). An initial report on mutations in the RNA splicing machinery found in MDS.
Papaemmanuil, E. et al. Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011). This study shows that mutations in the splicing machinery gene SF3B1 are associated with ringed sideroblasts in MDS.
Visconte, V. et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia 26, 542–545 (2012).
Makishima, H. et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 119, 3203–3210 (2012).
Graubert, T. A. et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nature Genet. 44, 53–57 (2011).
Damm, F. et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood 119, 3211–3218 (2012).
Patnaik, M. M. et al. SF3B1 mutations are prevalent in myelodysplastic syndromes with ring sideroblasts but do not hold independent prognostic value. Blood 119, 569–572 (2012).
Thol, F. et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 119, 3578–3584 (2012).
Haferlach, T. et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 28, 2529–2537 (2010).
Mills, K. I. et al. Microarray-based classifiers and prognosis models identify subgroups with distinct clinical outcomes and high risk of AML transformation of myelodysplastic syndrome. Blood 114, 1063–1072 (2009).
List, A. et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N. Engl. J. Med. 355, 1456–1465 (2006). The results of the clinical trial that led to the approval of lenalidomide for del(5q) MDS.
Raza, A. et al. Phase 2 study of lenalidomide in transfusion-dependent, low-risk, and intermediate-1 risk myelodysplastic syndromes with karyotypes other than deletion 5q. Blood 111, 86–93 (2008). This study shows that a percentage of patients with MDS without the del(5q) chromosomal deletion will respond to lenalidomide therapy.
Ebert, B. L. et al. An erythroid differentiation signature predicts response to lenalidomide in myelodysplastic syndrome. PLoS Med. 5, e35 (2008). A microarray analysis revealed an erythroid expression profile that may predict the response to lenalidomide in patients with non-del(5q) MDS.
Acknowledgements
The authors would like to thank C. Westbrook, D. Steensma and E. Estey for their critical reading of the manuscript and their helpful suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Cytopenias
-
Deficiencies of a cellular lineage of the blood.
- Hypolobated micromegakaryocytes
-
Abnormally small platelet-forming cells (megakaryocytes) with a decreased number of nuclear lobes.
- Ringed sideroblasts
-
Erythroblasts containing a ring of mitochondria filled with granules of ferritin around the nucleus.
- Thrombocytosis
-
High platelet counts in the blood.
- Spliceosome
-
A protein complex that is responsible for the excision of non-coding introns in precursor mRNA molecules.
Rights and permissions
About this article
Cite this article
Raza, A., Galili, N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat Rev Cancer 12, 849–859 (2012). https://doi.org/10.1038/nrc3321
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3321
This article is cited by
-
The evolution of preclinical models for myelodysplastic neoplasms
Leukemia (2024)
-
Caspase 8 deletion causes infection/inflammation-induced bone marrow failure and MDS-like disease in mice
Cell Death & Disease (2024)
-
Absence of early platelet increment in healthy mice during decitabine treatment
Scientific Reports (2022)
-
Predictive values of mutational variant allele frequency in overall survival and leukemic progression of myelodysplastic syndromes
Journal of Cancer Research and Clinical Oncology (2022)
-
Gene expression signatures associated with sensitivity to azacitidine in myelodysplastic syndromes
Scientific Reports (2020)