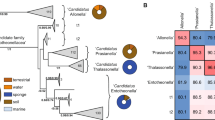
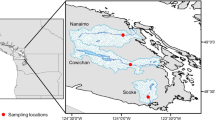
Abstract
Naturally produced polybrominated diphenyl ethers (PBDEs) pervade the marine environment and structurally resemble toxic man-made brominated flame retardants. PBDEs bioaccumulate in marine animals and are likely transferred to the human food chain. However, the biogenic basis for PBDE production in one of their most prolific sources, marine sponges of the order Dysideidae, remains unidentified. Here, we report the discovery of PBDE biosynthetic gene clusters within sponge-microbiome-associated cyanobacterial endosymbionts through the use of an unbiased metagenome-mining approach. Using expression of PBDE biosynthetic genes in heterologous cyanobacterial hosts, we correlate the structural diversity of naturally produced PBDEs to modifications within PBDE biosynthetic gene clusters in multiple sponge holobionts. Our results establish the genetic and molecular foundation for the production of PBDEs in one of the most abundant natural sources of these molecules, further setting the stage for a metagenomic-based inventory of other PBDE sources in the marine environment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Hites, R.A. Dioxins: an overview and history. Environ. Sci. Technol. 45, 16–20 (2011).
Wiseman, S.B. et al. Polybrominated diphenyl ethers and their hydroxylated/methoxylated analogs: environmental sources, metabolic relationships, and relative toxicities. Mar. Pollut. Bull. 63, 179–188 (2011).
Gribble, G.W. Naturally Occurring Organohalogen Compounds—A Comprehensive Update Vol. 91 (Springer, Vienna, 2010).
Agarwal, V. et al. Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat. Chem. Biol. 10, 640–647 (2014).
Sharma, G.M. & Vig, B. Studies on the antimicrobial substances of sponges. VI. Structures of two antibacterial substances isolated from the marine sponge Dysidea herbacea. Tetrahedr. Lett. 13, 1715–1718 (1972).
Carté, B. & Faulkner, D.J. Polybrominated diphenyl ethers from Dysidea herbacea, Dysidea chlorea and Phyllospongia foliascens. Tetrahedron 37, 2335–2339 (1981).
Calcul, L. et al. NMR strategy for unraveling structures of bioactive sponge-derived oxy-polyhalogenated diphenyl ethers. J. Nat. Prod. 72, 443–449 (2009).
Agarwal, V. et al. Complexity of naturally produced polybrominated diphenyl ethers revealed via mass spectrometry. Environ. Sci. Technol. 49, 1339–1346 (2015).
Unson, M.D., Holland, N.D. & Faulkner, D.J. A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar. Biol. 119, 1–11 (1994).
Becerro, M.A. & Paul, V.J. Effects of depth and light on secondary metabolites and cyanobacterial symbionts of the sponge Dysidea granulosa. Mar. Ecol. Prog. Ser. 280, 115–128 (2004).
Utkina, N.K. et al. Spongiadioxins A and B, two new polybrominated dibenzo-p-dioxins from an Australian marine sponge Dysidea dendyi. J. Nat. Prod. 64, 151–153 (2001).
Utkina, N.K., Denisenko, V.A., Virovaya, M.V., Scholokova, O.V. & Prokof'eva, N.G. Two new minor polybrominated dibenzo-p-dioxins from the marine sponge Dysidea dendyi. J. Nat. Prod. 65, 1213–1215 (2002).
Vetter, W. et al. Sponge halogenated natural products found at parts-per-million levels in marine mammals. Environ. Toxicol. Chem. 21, 2014–2019 (2002).
Teuten, E.L., Xu, L. & Reddy, C.M. Two abundant bioaccumulated halogenated compounds are natural products. Science 307, 917–920 (2005).
Shaul, N.J. et al. Nontargeted biomonitoring of halogenated organic compounds in two ecotypes of bottlenose dolphins (Tursiops truncatus) from the Southern California Bight. Environ. Sci. Technol. 49, 1328–1338 (2015).
Wan, Y. et al. Hydroxylated polybrominated diphenyl ethers and bisphenol A in pregnant women and their matching fetuses: placental transfer and potential risks. Environ. Sci. Technol. 44, 5233–5239 (2010).
Wang, H.S. et al. Hydroxylated and methoxylated polybrominated diphenyl ethers in blood plasma of humans in Hong Kong. Environ. Int. 47, 66–72 (2012).
Chen, A. et al. Hydroxylated polybrominated diphenyl ethers in paired maternal and cord sera. Environ. Sci. Technol. 47, 3902–3908 (2013).
Salomon, C.E. & Faulkner, D.J. Localization studies of ioactive cyclic peptides in the ascidian Lissoclinum patella. J. Nat. Prod. 65, 689–692 (2002).
Schmidt, E.W. et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 102, 7315–7320 (2005).
Hildebrand, M. et al. Approaches to identify, clone, and express symbiont bioactive metabolite genes. Nat. Prod. Rep. 21, 122–142 (2004).
Simmons, T.L. et al. Biosynthetic origin of natural products isolated from marine microorganism-invertebrate assemblages. Proc. Natl. Acad. Sci. USA 105, 4587–4594 (2008).
Anagnostidis, K. & Komárek, J. Modern approach to the classification system of cyanophytes. 3 - Oscillatoriales. Archiv für Hydrobiologie Suppl. 50–53, 327–472 (1988).
Hinde, R., Pironet, F. & Borowitzka, M.A. Isolation of Oscillatoria spongeliae, the filamentous cyanobacterial symbiont of the marine sponge Dysidea herbacea. Mar. Biol. 119, 99–104 (1994).
Ridley, C.P. et al. Speciation and biosynthetic variation in four dictyoceratid sponges and their cyanobacterial symbiont, Oscillatoria spongeliae. Chem. Biol. 12, 397–406 (2005).
Ridley, C.P., John Faulkner, D. & Haygood, M.G. Investigation of Oscillatoria spongeliae-dominated bacterial communities in four dictyoceratid sponges. Appl. Environ. Microbiol. 71, 7366–7375 (2005).
Medema, M.H. & Fischbach, M.A. Computational approaches to natural product discovery. Nat. Chem. Biol. 11, 639–648 (2015).
Wischang, D. & Hartung, J. Bromination of phenols in bromoperoxidase-catalyzed oxidations. Tetrahedron 68, 9456–9463 (2012).
Wischang, D., Radlow, M. & Hartung, J. Vanadate-dependent bromoperoxidases from Ascophyllum nodosum in the synthesis of brominated phenols and pyrroles. Dalton Trans. 42, 11926–11940 (2013).
Erpenbeck, D. et al. Evolution, radiation and chemotaxonomy of Lamellodysidea, a demosponge genus with anti-plasmodial metabolites. Mar. Biol. 159, 1119–1127 (2012).
Thacker, R.W. & Starnes, S. Host specificity of the symbiotic cyanobacterium Oscillatoria spongeliae in marine sponges, Dysidea spp. Mar. Biol. 142, 643–648 (2003).
Redmond, N.E. et al. Phylogeny and systematics of demospongiae in light of new small-subunit ribosomal DNA (18S) sequences. Integr. Comp. Biol. 53, 388–415 (2013).
Unson, M.D. et al. New polychlorinated amino acid derivatives from the marine sponge Dysidea herbacea. J. Org. Chem. 58, 6336–6343 (1993).
Hentschel, U. et al. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68, 4431–4440 (2002).
Hentschel, U., Piel, J., Degnan, S.M. & Taylor, M.W. Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 10, 641–654 (2012).
Agarwal, V. & Moore, B.S. Enzymatic synthesis of polybrominated dioxins from the marine environment. ACS Chem. Biol. 9, 1980–1984 (2014).
El Gamal, A. et al. Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair. Proc. Natl. Acad. Sci. USA 113, 3797–3802 (2016).
El Gamal, A., Agarwal, V., Rahman, I. & Moore, B.S. Enzymatic reductive dehalogenation controls the biosynthesis of marine bacterial pyrroles. J. Am. Chem. Soc. 138, 13167–13170 (2016).
Flatt, P. et al. Identification of the cellular site of polychlorinated peptide biosynthesis in the marine sponge Dysidea (Lamellodysidea) herbacea and symbiotic cyanobacterium Oscillatoria spongeliae by CARD-FISH analysis. Mar. Biol. 147, 761–774 (2005).
Fisch, K.M. et al. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat. Chem. Biol. 5, 494–501 (2009).
Podell, S. & Gaasterland, T. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol. 8, R16 (2007).
Fiedler, G., Arnold, M. & Maldener, I. Sequence and mutational analysis of the devBCA gene cluster encoding a putative ABC transporter in the cyanobacterium Anabaena variabilis ATCC 29413. Biochim. Biophys. Acta 1375, 140–143 (1998).
Taton, A. et al. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 42, e136 (2014).
Flórez, L.V., Biedermann, P.H., Engl, T. & Kaltenpoth, M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 32, 904–936 (2015).
Pennings, S.C., Pablo, S.R., Paul, V.J. & Duffy, J.E. Effects of sponge secondary metabolites in different diets on feeding by 3 groups of consumers. J. Exp. Mar. Biol. Ecol. 180, 137–149 (1994).
Kwan, J.C. et al. Genome streamlining and chemical defense in a coral reef symbiosis. Proc. Natl. Acad. Sci. USA 109, 20655–20660 (2012).
Marsh, G. et al. Identification, quantification, and synthesis of a novel dimethoxylated polybrominated biphenyl in marine mammals caught off the coast of Japan. Environ. Sci. Technol. 39, 8684–8690 (2005).
Malmvärn, A., Zebühr, Y., Kautsky, L., Bergman, K. & Asplund, L. Hydroxylated and methoxylated polybrominated diphenyl ethers and polybrominated dibenzo-p-dioxins in red alga and cyanobacteria living in the Baltic Sea. Chemosphere 72, 910–916 (2008).
Suyama, T.L., Cao, Z., Murray, T.F. & Gerwick, W.H. Ichthyotoxic brominated diphenyl ethers from a mixed assemblage of a red alga and cyanobacterium: structure clarification and biological properties. Toxicon 55, 204–210 (2010).
Schmidt, E.W. & Donia, M.S. Chapter 23. Cyanobactin ribosomally synthesized peptides--a case of deep metagenome mining. Methods Enzymol. 458, 575–596 (2009).
Nübel, U., Garcia-Pichel, F. & Muyzer, G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63, 3327–3332 (1997).
Parada, A.E., Needham, D.M. & Fuhrman, J.A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016).
Caporaso, J.G. et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267 (2010).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Paulson, J.N., Stine, O.C., Bravo, H.C. & Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202 (2013).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Podell, S. et al. Assembly-driven community genomics of a hypersaline microbial ecosystem. PLoS One 8, e61692 (2013).
Ma, A.T., Schmidt, C.M. & Golden, J.W. Regulation of gene expression in diverse cyanobacterial species by using theophylline-responsive riboswitches. Appl. Environ. Microbiol. 80, 6704–6713 (2014).
Clerico, E.M., Ditty, J.L. & Golden, S.S. Specialized techniques for site-directed mutagenesis in cyanobacteria. Methods Mol. Biol. 362, 155–171 (2007).
Acknowledgements
We thank our colleague B.M. Duggan at the University of California, San Diego for assistance in acquiring NMR data. This work was supported by the US National Science Foundation (DGE-1144086 Graduate Research Fellowship to J.M.B., OCE-1313747 to P.R.J., E.E.A., and B.S.M., IOS-1120113 to J.S.B., MCB-1149552 to E.E.A.); the US National Institutes of Health (K99ES026620 to V.A., R01-GM107557 to E.W.S., P01-ES021921 to P.R.J., E.E.A., and B.S.M., R01-CA172310 to V.J.P., instrument grant S10-OD010640); the US Department of Energy (DE-EE0003373 to J.W.G.); and the Helen Hay Whitney Foundation postdoctoral fellowship to V.A.
Author information
Authors and Affiliations
Contributions
V.A., S.P., A.T., E.W.S., V.J.P., E.E.A., and B.S.M. designed the study. V.A. performed chemical characterization; V.A., A.T., and J.W.G. performed cyanobacterial expression experiments; J.M.B., M.A.S., J.B., and P.R.J. performed phylogenetic analyses; S.P. performed metagenomic analyses; Z.L., V.J.P., and J.S.B. provided sponge samples, analytical tools and reagents; and V.A., J.M.B., S.P., E.E.A., and B.S.M. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–9 and Supplementary Figures 1–10 (PDF 2151 kb)
Supplementary Note
NMR Characterization Data (PDF 853 kb)
Rights and permissions
About this article
Cite this article
Agarwal, V., Blanton, J., Podell, S. et al. Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat Chem Biol 13, 537–543 (2017). https://doi.org/10.1038/nchembio.2330
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2330
This article is cited by
-
Testacosides A–D, glycoglycerolipids produced by Microbacterium testaceum isolated from Tedania brasiliensis
Applied Microbiology and Biotechnology (2024)
-
Sponges and their prokaryotic communities sampled from a remote karst ecosystem
Marine Biodiversity (2024)
-
The marine sponge genus Dysidea sp: the biological and chemical aspects—a review
Future Journal of Pharmaceutical Sciences (2023)
-
Minor Polybrominated Diphenyl Ethers from the Marine Sponge Dysidea fragilis
Chemistry of Natural Compounds (2023)
-
Synthase-selected sorting approach identifies a beta-lactone synthase in a nudibranch symbiotic bacterium
Microbiome (2023)