Abstract
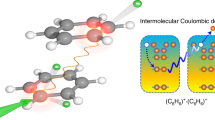
Proton transfer is ubiquitous in chemistry and biology, occurring, for example, in proteins, enzyme reactions and across proton channels and pumps. However, it has always been described in the context of hydrogen-bonding networks (‘proton wires’) acting as proton conduits. Here, we report efficient intramolecular ionization-induced proton transfer across a 1,3-dimethyluracil dimer, a model π-stacked system with no hydrogen bonds. Upon photoionization by tunable vacuum ultraviolet synchrotron radiation, the dimethyluracil dimer undergoes proton transfer and dissociates to produce a protonated monomer. Deuterated dimethyluracil experiments confirm that proton transfer occurs from the methyl groups and not from the aromatic C–H sites. Calculations reveal qualitative differences between the proton transfer reaction coordinate in the π-stacked and hydrogen-bonded base pairs, and that proton transfer in methylated dimers involves significant rearrangements of the two fragments, facilitating a relatively low potential energy barrier of only 0.6 eV in the ionized dimer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Schultz, T. et al. Efficient deactivation of a model base pair via excited-state hydrogen transfer. Science 306, 1765–1768 (2004).
Ghosh, A. K. & Schuster, G. B. Role of the guanine N1 imino proton in the migration and reaction of radical cations in DNA oligomers. J. Am. Chem. Soc. 128, 4172–4173 (2006).
Kennis, J. T. M. et al. Uncovering the hidden ground state of green fluorescent protein. Proc. Natl Acad. Sci. USA 101, 17988–17993 (2004).
Mitchell, P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148 (1961).
Chakraborty, T. in Charge Migration in DNA: Perspectives from Physics, Chemistry, and Biology (Springer Verlag, 2007).
Decoursey, T. E. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 83, 475–579 (2003).
Nagle, J. F. & Morowitz, H. J. Molecular mechanims for proton transport in membranes. Proc. Natl Acad. Sci. USA 75, 298–302 (1978).
Marx, D., Tuckerman, M. E., Hutter, J. & Parrinello, M. The nature of the hydrated excess proton in water. Nature 397, 601–604 (1999).
Sobolewski, A. L., Domcke, W. & Hattig, C. Tautomeric selectivity of the excited-state lifetime of guanine/cytosine base pairs: the role of electron-driven proton-transfer processes. Proc. Natl Acad. Sci. USA 102, 17903–17906 (2005).
Perun, S., Sobolewski, A. L. & Domcke, W. Role of electron-driven proton-transfer processes in the excited-state deactivation adenine–thymine base pair. J. Phys. Chem. A 110, 9031–9038 (2006).
Wahl, M. C. & Sundaralingam, M. C–H···O hydrogen bonding in biology. Trends Biochem. Sci. 22, 97–102 (1997).
Marfurt, J. & Leumann, C. Evidence for C–H=O hydrogen bond assisted recognition of a pyrimidine base in the parallel DNA triple-helical motif. Angew. Chem. Int. Ed. 37, 175–177 (1998).
Loganathan, D. & Aich, U. Observation of a unique pattern of bifurcated hydrogen bonds in the crystal structures of the N-glycoprotein linkage region models. Glycobiology 16, 343–348 (2006).
Somorjai, G. A., Tao, F. & Butcher, D. in Scanning Tunneling Microscopy in Surface Science 189–217 (Wiley-VCH Verlag, 2010).
Keiluweit, M. & Kleber, M. Molecular-level interactions in soils and sediments: the role of aromatic π-systems. Environ. Sci. Technol. 43, 3421–3429 (2009).
Crespo-Hernandez, C. E., Cohen, B., Hare, P. M. & Kohler, B. Ultrafast excited-state dynamics in nucleic acids. Chem. Rev. 104, 1977–2019 (2004).
Middleton, C. T. et al. DNA excited-state dynamics: from single bases to the double helix. Annu. Rev. Phys. Chem. 60, 217–239 (2009).
Kanvah, S. et al. Oxidation of DNA: damage to nucleobases. Acc. Chem. Res. 43, 280–287 (2009).
Schreier, W. J. et al. Thymine dimerization in DNA is an ultrafast photoreaction. Science 315, 625–629 (2007).
Bhosale, S. et al. Photoproduction of proton gradients with π-stacked fluorophore scaffolds in lipid bilayers. Science 313, 84–86 (2006).
Sisson, A. L., Shah, M. R., Bhosale, S. & Matile, S. Synthetic ion channels and pores (2004–2005). Chem. Soc. Rev. 35, 1269–1286 (2006).
Satzger, H., Townsend, D. & Stolow, A. Reassignment of the low lying cationic states in gas phase adenine and 9-methyl adenine. Chem. Phys. Lett. 430, 144–148 (2006).
Meredith, P. & Sarna, T. The physical and chemical properties of eumelanin. Pigment Cell Res. 19, 572–594 (2006).
d'Ischia, M., Napolitano, A., Pezzella, A., Meredith, P. & Sarna, T. Chemical and structural diversity in eumelanins: unexplored bio-optoelectronic materials. Angew. Chem. Int. Ed. 48, 3914–3921 (2009).
Kobayashi, K. & Tagawa, S. Direct observation of guanine radical cation deprotonation in duplex DNA using pulse radiolysis. J. Am. Chem. Soc. 125, 10213–10218 (2003).
Adhikary, A., Khanduri, D. & Sevilla, M. D. Direct observation of the hole protonation state and hole localization site in DNA-oligomers. J. Am. Chem. Soc. 131, 8614–8619 (2009).
Gador, N. et al. Electronic structure of adenine and thymine base pairs studied by femtosecond electron-ion coincidence spectroscopy. J. Phys. Chem. A 111, 11743–11749 (2007).
Nir, E., Plutzer, C., Kleinermanns, K. & de Vries, M. Properties of isolated DNA bases, base pairs and nucleosides examined by laser spectroscopy. Eur. Phys. J. D 20, 317–329 (2002).
Plutzer, C., Hunig, I. & Kleinermanns, K. Pairing of the nucleobase adenine studied by IR–UV double-resonance spectroscopy and ab initio calculations. Phys. Chem. Chem. Phys. 5, 1158–1163 (2003).
Zadorozhnaya, A. A. & Krylov, A. I. Ionization-induced structural changes in uracil dinners and their spectroscopic signatures. J. Chem. Theoret. Comput. 6, 705–717 (2010).
Zadorozhnaya, A. A. & Krylov, A. I. Zooming into π-stacked manifolds of nucleobases: ionized states of dimethylated uracil dimers. J. Phys. Chem. A 114, 2001–2009 (2010).
Golubeva, A. A. & Krylov, A. I. The effect of π-stacking and H-bonding on ionization energies of a nucleobase:uracil dimer cation. Phys. Chem. Chem. Phys. 11, 1303–1311 (2009).
Bravaya, K. B., Kostko, O., Ahmed, M. & Krylov, A. I. The effect of π-stacking, H-bonding, and electrostatic interactions on the ionization energies of nucleic acid bases: adenine–adenine, thymine–thymine and adenine–thymine dimers. Phys. Chem. Chem. Phys. 12, 2292–2307 (2010).
de Vries, M. S. & Hobza, P. Gas-phase spectroscopy of biomolecular building blocks. Annu. Rev. Phys. Chem. 58, 585–612 (2007).
Munson, M. S. B. & Field, F. H. Chemical ionization mass spectrometry. I. General introduction. J. Am. Chem. Soc. 88, 2621–2630 (1966).
Gronert, S., Feng, W. Y., Chew, F. & Wu, W. The gas phase acid/base properties of 1,3,-dimethyluracil, 1-methyl-2-pyridone, and 1-methyl-4-pyridone: relevance to the mechanism of orotidine-5′-monophosphate decarboxylase. Int. J. Mass Spectrom. 195–196, 251–258 (2000).
Jochims, H. W., Schwell, M., Baumgartel, H. & Leach, S. Photoion mass spectrometry of adenine, thymine and uracil in the 6–22 eV photon energy range. Chem. Phys. 314, 263–282 (2005).
Kabelac, M. & Hobza, P. At nonzero temperatures, stacked structures of methylated nucleic acid base pairs and microhydrated nonmethylated nucleic acid base pairs are favored over planar hydrogen-bonded structures: a molecular dynamics simulations study. Chem. Euro. J. 7, 2067–2074 (2001).
Kabelac, M. & Hobza, P. Potential energy and free energy surfaces of all ten canonical and methylated nucleic acid base pairs: molecular dynamics and quantum chemical ab initio studies. J. Phys. Chem. B 105, 5804–5817 (2001).
He, Y. G., Wu, C. Y. & Kong, W. Photophysics of methyl-substituted uracils and thymines and their water complexes in the gas phase. J. Phys. Chem. A 108, 943–949 (2004).
Boerjan, W., Ralph, J. & Baucher, M. Lignin biosynthesis. Ann. Rev. Plant Biol. 54, 519–546 (2003).
Takahashi, L. K. et al. VUV photoionization and mass spectrometric characterization of the lignin monomers coniferyl and sinapyl alcohol. J. Phys. Chem. A 115, 3279–3290 (2011).
Belau, L., Wilson, K. R., Leone, S. R. & Ahmed, M. Vacuum–ultraviolet photoionization studies of the microhydration of DNA bases (guanine, cytosine, adenine, and thymine). J. Phys. Chem. A 111, 7562–7568 (2007).
Shao, Y. et al. Advances in methods and algorithms in a modern quantum chemistry program package. Phys. Chem. Chem. Phys. 8, 3172–3191 (2006).
Acknowledgements
Experiments were carried out at the Advanced Light Source, and the d6-1,3-mU was synthesized at the Molecular Foundry, both at Lawrence Berkeley National Laboratory. Berkeley participants are supported by the Office of Science, Office of Basic Energy Sciences, of the US Department of Energy (contract no. DE-AC02-05CH11231), through the Chemical Sciences Division (A.G., O.K., S.R.L., M.A.) and the Materials Sciences Division (R.K.). R.K. is also supported by the Defense Threat Reduction Agency (IACRO-B0845281). This work was conducted in the framework of the iOpenShell Center (iopenshell.usc.edu), supported by the National Science Foundation through CRIF:CRF (CHE-0625419 + 0624602 + 0625237 and CHE-0951634, to A.I.K.) grants.
Author information
Authors and Affiliations
Contributions
M.A. conceived and designed the experiments. A.G. and O.K. conducted the experiments. R.K. synthesized the deuterated compounds. K.B.B. and A.I.K. performed electronic structure calculations. A.G. and K.B.B. contributed equally to this work. A.G., K.B.B., A.I.K., M.A. and S.R.L. co-wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 1712 kb)
Rights and permissions
About this article
Cite this article
Golan, A., Bravaya, K., Kudirka, R. et al. Ionization of dimethyluracil dimers leads to facile proton transfer in the absence of hydrogen bonds. Nature Chem 4, 323–329 (2012). https://doi.org/10.1038/nchem.1298
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1298
This article is cited by
-
Water acting as a catalyst for electron-driven molecular break-up of tetrahydrofuran
Nature Communications (2020)
-
C70 Fullerene Cage as a Novel Catalyst for Efficient Proton Transfer Reactions between Small Molecules: A Theoretical study
Scientific Reports (2019)
-
Effects of single water molecule on proton transfer reaction in uracil dimer cation
Theoretical Chemistry Accounts (2016)
-
A TDDFT Study on the Excited-State Intramolecular Proton Transfer (ESIPT): Excited-State Equilibrium Induced by Electron Density Swing
Journal of Fluorescence (2013)