Abstract
Influenza A viruses are important human pathogens causing periodic pandemic threats. Nonstructural protein 1 (NS1) protein of influenza A virus (NS1A) shields the virus against host defense. Here, we report the crystal structure of NS1A RNA-binding domain (RBD) bound to a double-stranded RNA (dsRNA) at 1.7Å. NS1A RBD forms a homodimer to recognize the major groove of A-form dsRNA in a length-independent mode by its conserved concave surface formed by dimeric anti-parallel α-helices. dsRNA is anchored by a pair of invariable arginines (Arg38) from both monomers by extensive hydrogen bonds. In accordance with the structural observation, isothermal titration calorimetry assay shows that the unique Arg38-Arg38 pair and two Arg35-Arg46 pairs are crucial for dsRNA binding, and that Ser42 and Thr49 are also important for dsRNA binding. Agrobacterium co-infiltration assay further supports that the unique Arg38 pair plays important roles in dsRNA binding in vivo.
Similar content being viewed by others
Introduction
Influenza viruses are one of the lethal viruses threatening human health, and cause pandemic spread almost once every 10 years – e.g. the pandemic spread in 1918 that resulted in 30 million deaths 1. The recent outbreak of avian H5N1 viruses in birds has the potential to cause the next round of pandemic spread in humans.
The influenza viruses are RNA viruses belonging to Orthomyxoviridae family. Influenza viruses are of three types, of which type A is the most virulent human pathogen and causes the most severe disease. The influenza A genome contains eight pieces of segmented negative-sense RNAs and encodes 11 proteins, including the nonstructural protein 1 (NS1) 2.
Influenza A NS1 protein (NS1A) was reported to have multiple functions in modulating immune responses. NS1A is a viral immuno-suppressor which inhibits innate immunity by preventing type I interferon (IFN) release and inhibits adaptive immunity by attenuating human dendritic cell (DC) maturation and the capacity of DCs to induce T-cell responses 3. NS1A accumulates in the nuclei of host cells through binding to importin α by two nuclear localization signal (NLS) sequences, NLS1 (residues 35-41) and NLS2 (residues 231-237) 4. NS1A was further reported to suppress the antiviral host defense at multiple levels by blocking both the activation of IFN regulatory factor 3 and post-transcriptional processing of cellular mRNAs 5. NS1A regulates a major component of the cellular antiviral system, PKR, while activated PKR inhibits viral and cellular protein synthesis and virus replication by targeting the translation initiation factor eIF2. NS1A protein counteracts PKR-mediated antiviral response through the direct interaction with PKR by a five-residue short peptide sequence (123IMDKN127) 6, 7.
NS1A is also an RNA-binding protein; it was reported as a poly(A)-binding protein that inhibits the nuclear export of mRNAs 8, 9 and is involved in modulating pre-mRNA splicing both in vitro and in vivo 10. Importantly, the biological functions of NS1A in immune response probably involve its double-stranded RNA (dsRNA)-binding ability. NS1A protein encoded by influenza virus was proposed to suppress antiviral response by shielding the virus 11. dsRNA binding by NS1A proteins is important to protect the viruses against human antiviral response during infection 12. One of the primary functions of RNA binding by NS1A in infected cells is sequestering dsRNA from the 2′-5′ oligo (A) synthetase/ribonuclease L pathway 12.
NS1A protein has an N-terminal RNA-binding domain (RBD) and a C-terminal effector domain, and the structures of these domains have been determined in the free state 13, 14, 15, 16. Previously, the crystal structures of NS1A RBD and NS1B RBD revealed that NS1 RBD domain forms a homodimeric six-helical fold for dsRNA recognition 14, 16. The NMR chemical shift perturbation in NS1A RBD upon complex formation with dsRNA was used to predict the protein-dsRNA interactions and the potential dsRNA recognition modes 16. Although the model derived from the NMR chemical shift perturbation provides a visualized template for further experimental designs, the molecular details of dsRNA recognition by NS1A RBD are still unclear.
To study the structural basis of dsRNA recognition by NS1A and experimentally validate the major features proposed by previous reports, we have determined here the crystal structure of the RBD of NS1A bound to a self-complementary 21-nucleotide siRNA (with 19 base pairs) at 1.7Å (Figure 1A); the crystallographic data and statistics are summarized in Table 1. Our complex structure reveals that NS1A RBD recognizes dsRNA in a major groove binding mode and the recognition is entirely with dsRNA backbone. Structural and biochemical analyses further address the importance of the unique Arg38-Arg38 pair and Arg35-Arg46 pairs for dsRNA binding.
Overall structure of NS1A RBD bound to siRNA duplex. (A) Nucleic acid sequence of siRNA duplex used for co-crystallization. (B) Ribbon representation of NS1A RBD-siRNA duplex complex showing that the NS1A RBD dimer recognizes the major groove of RNA duplex, with a pair of invariable Arg38 serving as anchor residues. One monomer of NS1A RBD is colored in green and the other is colored in cyan, while the RNA duplex is colored in orange. Key residues involved in RNA duplex recognition are colored in magenta (direct hydrogen bond interaction), blue (hydrogen bond interaction via water bridges), wheat (electrostatic interaction) and red (anchoring residues), respectively. (C) Rotation of (B) along Y-axis by 90°. The α-helices (α2 and α2′) providing the concave surface for RNA duplex recognition are indicated. The Arg38 penetrating into the helical turn of RNA duplex for anchoring is also indicated. (D) Rotation of (B) along X-axis by 90°. Secondary structures of NS1A RBD are indicated. (E) Rotation of (B) along Y-axis by 180°. Secondary structures of NS1A RBD are indicated.
Results
Crystal structure of the complex
NS1A RBD domain forms a dimeric six-helical fold harboring a shallow concave surface that recognizes the major groove of the dsRNA. The dsRNA duplex helical axis is approximately parallel in relation to the axes of the anti-parallel helices α2 and α2′ (Figure 1B-1E). The RNA helix adopts an A-form conformation without significant bending along the helical axis at the middle part; however, it has ∼40° bending towards the protein at both ends of the helical turn to facilitate the RNA-protein interactions (Figure 1C and 1D). The electron density at the 3′-dinucleotide overhang of the bound siRNA is weak and probably disordered in the crystal structure. There is a deep pocket underneath the dsRNA-binding surface, with the wall formed by anti-parallel helices 2 and 2′, the base formed by anti-parallel helices 1 and 1′, and the lid formed by the invariable residues Arg38 from both monomers (Figure 1B; Supplementary information, Figure S1A).
Key residues involved in dsRNA recognition
The NS1A RBD domain dimer buries 1 978Å2 of total solvent-accessible area of the bound siRNA duplex. The positively charged residues, such as Arg35, Arg37, Arg38 and Lys41, are clustered at the middle of the dsRNA-binding surface and play primary roles in making hydrogen bonds and electrostatic interactions with both strands of the dsRNA (Supplementary information, Figure S1A). Among them, the invariable residue Arg38 penetrates into dsRNA helix and forms hydrogen bonds simultaneously with the phosphorus of two nucleotides (C7-O1P and C7-O2P) from two different strands (Figure 1B and 1C; Supplementary information, Figures S1A and S1B; Table 2). Furthermore, Arg38 forms hydrogen bonds with its symmetric related molecule to form a hydrogen-bond network to anchor the dsRNA and prevent it from sliding along the binding surface (Figure 1B and 1C; Supplementary information, Figure S1A). Another two pairs of hydrogen bonds formed between Arg35 from one monomer and Arg46 from the symmetric related molecule and paralleling to the Arg38 pair are also involved in dsRNA binding through the hydrogen bonding between Arg35 and the bound dsRNA (Figure 2A and Table 2).
Structural comparison of NS1A-RBD dimer in dsRNA bound state and in free state. (A) NS1A-RBD dimer in dsRNA bound state. Arg38 pair and Arg35-Arg46 pairs, which are critical for dsRNA binding, are indicated and colored in red. The bound glycerol molecules are colored in wheat. The omit maps on residues Arg38 and glycerol were calculated and contoured at 4.5σ. (B) NS1A-RBD dimer in free state. The critical Arg38 pair for dsRNA anchoring is unpaired in free state. (C) Electrostatic potential surface view of (A). Conserved positively charged residues clustered at the middle of RNA duplex binding surface for recognition through hydrogen bonds or electrostatic interactions are indicated. (D) Electrostatic potential surface view of (B).
Several other residues outside this positive patch, such as Thr5, Asp29, Asp34, Ser42 and Thr49, also contribute to the dsRNA binding by forming hydrogen bonds with dsRNA either directly or via water bridges (Supplementary information, Figure S1C and Table 2). In accordance with the structural observation, isothermal titration calorimetry (ITC) assay showed that mutations on the key residues involved in direct interactions with dsRNA have significant impacts on dsRNA binding. Both S42A and T49A decrease the dsRNA-binding affinity by 10-fold (Figure 3B and 3C), whereas R35AR46A double mutations or R38A single mutation abolishes dsRNA binding (Figure 3G and 3H) as indicated by the similar binding curves to that of the negative control (Figure 3F). The ITC data are consistent with the previous report that Arg38 is the only amino acid absolutely required for dsRNA binding 17.
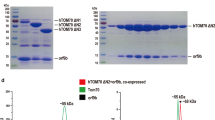
Characterization of key residues involved in dsRNA binding. (A-H) ITC data of NS1A and its mutants binding with the 21-nt siRNA duplex. Raw titration data and integrated heat measurements are shown in the upper and lower plots, respectively. The Ka and stoichiometry numbers (n) obtained by fitting a standard one-interaction-site model are reported, with the associated s.d. determined by nonlinear least-squares analysis. (I) Suppression of RNA silencing in N. benthamiana by NS1A RBD. Wild-type NS1A RBD (lower left spot on the leaf) and the R38A mutant (lower right spot on the leaf) expressed in Agrobacterium tumefaciens were co-infiltrated with the GFP expression vector into GFP-expressing N. benthamiana leafs (C16), respectively. Infiltration of the GFP expression vector alone served as a negative control (upper left spot on the leaf), while co-infiltration of the TBSV p19 served as a positive control (upper right spot on the leaf).
NS1A RBD has the ability to distinguish between dsRNA and dsDNA and recognizes dsRNA in a sequence-independent manner because all the intermolecular contacts are directed towards the sugar-phosphate backbone and 2′-OH groups on the RNA strand. Arg35 and Thr49 directly recognize 2′-OH groups of RNA, whereas Thr5, Asp29 and Asp34 recognize 2′-OH groups of RNA via water bridges (Supplementary information, Figure S1C and Table 2).
NS1A RBD suppresses RNA silencing in vitro
NS1 protein was reported to have an RNA-silencing suppression activity in plants 18, 19, similar to established RNA-silencing suppressor proteins, such as cucumovirus 2b (CMV2b) and flock house virus B2 (FHVB2) 20, 21. To test whether NS1A RBD domain itself is sufficient to suppress RNAi in vitro and whether Arg38 is responsible for dsRNA binding in vivo, we performed Agrobacterium co-infiltration assay in transgenic plants that express green fluorescent protein (GFP), as described previously 22. In this transgenic plant, transient introduction of GFP via Agrobacterium infiltration results in GFP silencing, while an RNA silencing suppressor can rescue GFP expression in the infiltrated regions.
Our results demonstrate that transient expression of NS1A RBD suppressed RNA-mediated silencing of the introduced GFP gene, leading to a strong green fluorescence when examined under ultraviolet (UV) illumination (Figure 3I, lower left spot on the leaf), similar to the suppression by the P19 protein (Figure 3I, upper right spot on the leaf). As a control, GFP protein expression was almost completely silenced when the GFP expression vector was infiltrated alone (Figure 3I, upper left spot on the leaf).
By contrast, a single mutation of Arg38 (R38A) led to a significant decrease of GFP expression (Figure 3I, lower right spot on the leaf), suggesting that the ability of NS1A RBD to suppress RNA-mediated silencing is abolished by the point mutation. The correlation between RNA-binding affinity and the ability to suppress RNA silencing supports a potential role of NS1 in counteracting innate antiviral response in vertebrates through sequestering RNA duplexes.
Discussion
Structural roles of Arg35-Arg46 and Arg38-Arg38 pairs
Our structural and functional analyses revealed that several key residues on the concave dsRNA-binding surface, such as Arg35, Arg38, Ser42 and Thr49, are directly involved in dsRNA binding and the corresponding mutants showed significantly reduced binding affinity with the 21-nt siRNA duplex (Figure 3 and Table 2). Importantly, dsRNA binding is abolished for the R38A single mutant and R35AR46A double mutant. The Arg38-Arg38 pair and Arg35-Arg46 pairs are located at the concave surface and are crucial for dsRNA binding, with the former penetrating into the bound dsRNA and the latter probably critical for the stabilization of NS1A RBD dimer (Figure 2A and 2C).
Notably, there is a dramatic conformational change in the orientation of the side chain of Arg38 before and after dsRNA binding, whereas the overall structure and the orientation of other residues are almost identical (root mean square deviation (r.m.s.d.) 0.453Å, 140 Cα) (Figure 2A-2D). The Arg38 pair acts as a lid to cover a deep pocket underneath the concave dsRNA-binding surface (Figure 2A and 2C). The binding of dsRNA triggers the Arg38 pair to form a lid covering the deep pocket (Figure 2A and 2C), whereas the release of dsRNA induces the breakage of Arg38 pair to open the deep pocket (Figure 2B and 2D).
Potential drug-binding pocket
The conserved deep pocket observed underneath the concave dsRNA-binding surface was suggested as an inviting target for structure-function studies and antiviral drug design 16. Notably, we have observed large extra densities at this deep pocket, which were assigned as glycerol molecules (Figure 2A). The chemical compound binding site identified at this pocket underneath the invariable Arg38 pair from the crystal structure strongly suggests that this pocket could be a novel drug target.
Correlation between the dsRNA-binding affinity of NS1 proteins and the virulence of influenza viruses is documented in the literature. Any efforts resulting in the disruption of Arg38 pair could lead to the decrease of dsRNA-binding affinity by NS1 and could eventually lead to the attenuation of virulence. The observation of the chemical compound binding site at this pocket suggests that some chemical compounds might have the abilities to bind to this pocket and induce the disruption of Arg38 pair. Such compounds would be candidates for developing novel antiviral drugs.
Earlier predictions of dsRNA recognition by NS1A RBD
Several predictions have been made regarding NS1A RBD recognition of dsRNA based on the crystal structure of NS1A RBD and NS1B RBD in free form and NMR chemical shift perturbation upon complex formation between NS1A RBD and dsRNA 13, 14, 15, 16. It was suggested from the free NS1A-RBD structure that residues Phe14, Lys20, Arg37, Arg38, Lys41, Arg44, Lys62, Lys67 and Lys70 are involved in dsRNA binding 14. Previous biochemical analysis suggested that the residues critical for dimer formation, such as Asp12, Arg19, Asp29, Arg35 and Arg46, are important for dsRNA binding 17. Although Arg37, Lys41 and Arg44 on the dsRNA-binding surface were proposed to have important roles in dsRNA binding, the only residue absolutely required for dsRNA binding was shown to be Arg38 17.
NMR chemical shift perturbation upon complex formation indicated that Thr5, Pro31, Asp34, Arg35, Arg38, Lys41, Gly45, Arg46 and Thr49 are involved in dsRNA binding 16. Although the major groove binding mode was preferred by NMR perturbation analysis, the authors could not unambiguously distinguish the two potential binding modes (major groove versus minor groove) nor rule out the possibility of rapid inter-conversion between the two different modes.
Our structure here addresses a long-standing challenge associated with the molecular basis of recognition and shielding of the viral dsRNA by NS1A RBD. Our structural and functional analyses indicate that, although all the residues involved in dsRNA interactions are highly conserved among NS1A proteins of all influenza A viruses, some conserved residues, such as Pro31, Gly45 and Arg46, are not directly involved in dsRNA binding, which is different from the previous notion suggested by NMR data 16. Although both Arg46 and Thr49 were previously proposed to interact with the 2′-OH group of RNA 16, we only observed the direct interaction between Thr49 and the 2′-OH group (Supplementary information, Figure S1C and Table 2). No direct interactions are observed between Arg46 and the 2′-OH group. However, Arg46 is indirectly involved in dsRNA binding through hydrogen bonding with Arg35, which directly interacts with the bound dsRNA. Our complex structure further shows that residues Phe14, Lys20, Arg37, Arg44, Lys62, Lys67 and Lys70, which were proposed to interact with dsRNA based on the free NS1A-RBD structure 14, are in fact not involved in dsRNA binding. The notion that Lys20 and Arg44 are not required for dsRNA binding is consistent with the previously published results 17. In accordance with the structural observation, our ITC binding assay indicated that the dsRNA-binding affinity of the R37A mutant is similar to that of wild type, whereas that of the R44A mutant is only modestly decreased (Figure 3D and 3E).
Comparison with other RNA-silencing suppressors
NS1A RBD shares overall structural similarity with FHVB2 (N-terminal 1-73) in dsRNA recognition; however, there are also significant structural differences between them. Firstly, although both NS1A RBD and FHVB2 (N-terminal 1-73) recognize dsRNA via the platform formed by the anti-parallel dimeric helices α2 and α2′ in a size- and sequence-independent manner, NS1A RBD recognizes the major groove of the dsRNA (Figure 4A), whereas FHVB2 dimer recognizes two adjacent minor grooves on one face of the dsRNA 23 (Figure 4B). Secondly, the long axis of NS1A RBD dimer is rotated about 40° in relation to the RNA helical axis (Figure 1B), whereas the long axis of the FHVB2 dimer, which has 10 more amino acids than that of NS1A RBD, is rotated about 10° in relation to the RNA helical axis. Thirdly, dsRNA within the NS1A RBD complex has ∼40° bending at the end of its helical turn to facilitate the RNA-protein binding (Figure 4A), whereas dsRNA bound on FHVB2 has no significant helical bending (Figure 4B). The significant bending of dsRNA suggests that NS1A dimer might adopt a different binding mode to bind long dsRNA in vivo, distinct from the mechanism proposed by the FHVB2 structure 23.
Materials and Methods
Protein expression and purification
Recombinant NS1A dsRNA-binding domain (RBD, residue 1-70) was generated by cloning the open reading frame into EcoRI and NotI sites of pGEX 6P-1 (Ampicillin selection, Novagen, www.novagen.com). NS1A RBD mutants were prepared using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, www.stratagene.com), and the constructs were verified by sequencing. To express recombinant proteins, transformed BL21(DE3)/RILP Escherichia coli were grown to an OD600 ∼0.6 and then protein expression was induced overnight at 20 °C using 0.4 mM isopropyl β-D-thiogalactoside (IPTG). Cells were harvested by centrifugation.
For purification, cells were re-suspended in 1× PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3), complete proteinase inhibitor (Roche, www.roche.com), and lysed by a cell disruptor. After centrifugation (40 000 × g, 1 h), the supernatant was loaded onto a glutathione sepharose affinity column equilibrated in 1× PBS. Non-specific binding proteins were washed out by 1× PBS buffer and the fused protein was eluted by elution buffer (0.154 g reduced glutathione dissolved in 50 ml 50 mM Tris-HCl, pH 8.0). Pooled fractions from the glutathione sepharose affinity column were concentrated and the N-terminal GST-tag was cleaved by PreScission protease before loading on a HiLoad Superdex G-75 column (GE health care) equilibrated in 500 mM NaCl, 10 mM DTT and 25 mM Tris, pH 7.4 for further purification. The peak fractions from the Superdex G-75 were pooled together and concentrated to about 10 ml, then purified through Superdex G-75 again. The peak fractions were pooled together and concentrated to 75 mg/ml in a Microcon (Amicon).
Protein crystallization, data collection and structure determination
For NS1A RBD/RNA duplex complex crystallization, 10 μl 75 mg/ml NS1A RBD (in 500 mM NaCl) was mixed with 2 mM 52 μl 21-nucleotide RNA dissolved in water (NS1A RBD:siRNA21=1:1.1 in molar ratio). The final concentration of NS1A RBD within the complex was about 12 mg/ml. Crystals of complex were grown by hanging drop vapor diffusion at 20 °C. Typically, a 2.0-μl hanging drop contained 1.0 μl of protein/RNA complex mixed with 1.0 μl of reservoir solution containing 0.02 M magnesium acetate, 0.05 M 4-morpholineethanesulfonic acid (MES) (pH 5.2) and 2.6 M ammonium sulfate, with 10 mM adenosine-5-triphosphate disodium salt as additive, and equilibrated over 1 ml of reservoir solution. These crystals grew as a cluster to a maximum size of 0.1 mm×0.05 mm×0.05 mm over the course of 7 days.
For data collection, crystals were flash frozen (100 K) in the above reservoir solution supplemented with 30% glycerol. A total of 360 frames of 1° oscillation were collected for each crystal on beamline X12C at the National Synchrotron Light Source at Brookhaven National Laboratory and was processed by HKL2000 (www.hkl-xray.com). The crystals belong to space group C2, with unit cell parameters listed in Table 1.
The crystal structure of NS1A RBD/RNA complex was determined by molecular replacement using NS1A RBD monomer as a search model (PDBID: 1AIL). There are two molecules at one asymmetric unit. The siRNA duplex was built at the late stage of the refinement without ambiguity due to the high resolution of the structure. The model was further rebuilt by using the program O 24 and refined using REFMAC/CCP4 25. The R-free set contained 5% of the reflections chosen at random. The model comprises residues 1-70 of the protein and nucleotides 1-21 of the RNA.
Isothermal titration calorimetry assay
All experiments were performed in a buffer containing 25 mM Tris-HCl (pH 7.4) and 50 mM NaCl. Protein (10-15 μM) and RNA (200 μM) samples were filtered and degassed before titration. Protein samples were loaded into the cell and RNA samples were loaded into the syringe with a stirring speed of 310 rpm (Microcal VP-ITC calorimeter, Northampton, MA). Data were collected in high feedback mode with a filter period of 2 s. The calorimetric data were processed and fitted into the single set of identical sites model using Microcal Origin (Version 5.0) and analyzed by the software supplied by the instrument.
Agrobacterium co-infiltration assay
NS1A RBD domain and R38A mutant were cloned into the pBA vector to generate constructs for infiltration as described previously 22. These plasmids were then transformed into Agrobacterium tumefaciens strain EHA105 by electroporation and infiltrated into transgenic Nicotiana benthamiana expressing the GFP transgene (line 16C). The GFP fluorescence expression at N. benthamiana leaf was visualized 6 days after infiltration by a long-wave UV lamp (SB-100P/F High intensity Ultraviolet lamp, Spectronics, USA, www.spetroline.com) and photographed by a Nikon D-80 digital camera with a yellow filter.
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Abbreviations
- NS1A:
-
(NS1 protein of influenza A virus)
- dsRNA:
-
(double-stranded RNA)
- RBD:
-
(RNA-binding domain)
- MES:
-
(4-morpholineethanesulfonic acid)
- r.m.s.d.:
-
(root mean square deviation)
- ITC:
-
(isothermal titration calorimetry)
References
Reid AH, Taubenberger JK, Fanning TG . The 1918 Spanish influenza: integrating history and biology. Microbes Infect 2001; 3:81–87.
Ghedin E, Sengamalay N, Shumway M, et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 2005; 437:1162–1166.
Fernandez-Sesma A, Marukian S, Ebersole BJ, et al. Influenza virus evades innate and adaptive immunity via the NS1 protein. J Virol 2006; 80:6295–6304.
Melen K, Kinnunen L, Fagerlund R, et al. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J Virol 2007; 81:5995–6006.
Kochs G, Garcia-Sastre A, Martinez-Sobrido L . Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 2007; 81:7011–7021.
Bergmann M, Garcia-Sastre A, Carnero E, et al. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J Virol 2000; 74:6203–6206.
Min JY, Li S, Sen GC, Krug RM . A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 2007; 363:236–243.
Qiu Y, Krug RM . The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J Virol 1994; 68:2425–2432.
Lu Y, Wambach M, Katze MG, Krug RM . Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 1995; 214:222–228.
Qiu Y, Nemeroff M, Krug RM . The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA 1995; 1:304–316.
Newby CM, Sabin L, Pekosz A . The RNA binding domain of influenza A virus NS1 protein affects secretion of tumor necrosis factor alpha, interleukin-6, and interferon in primary murine tracheal epithelial cells. J Virol 2007; 81:9469–9480.
Min JY, Krug RM . The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′–5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci USA 2006; 103:7100–7105.
Chien CY, Tejero R, Huang Y, et al. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat Struct Biol 1997; 4:891–895.
Liu J, Lynch PA, Chien CY, Montelione GT, Krug RM, Berman HM . Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol 1997; 4:896–899.
Bornholdt ZA, Prasad BV . X-ray structure of influenza virus NS1 effector domain. Nat Struct Mol Biol 2006; 13:559–560.
Yin C, Khan JA, Swapna GV, et al. Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J Biol Chem 2007; 282:20584–20592.
Wang W, Riedel K, Lynch P, Chien CY, Montelione GT, Krug RM . RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 1999; 5:195–205.
Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M . The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol 2004; 85:983–991.
Delgadillo MO, Sáenz P, Salvador B, García JA, Simón-Mateo C . Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J Gen Virol 2004; 85:993–999.
Zhang X, Yuan YR, Pei Y, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 2006; 20:3255–3268.
Li H, Li WX, Ding SW . Induction and suppression of RNA silencing by an animal virus. Science 2002; 296:1319–1321.
Voinnet O, Lederer C, Baulcombe DC . A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 2000; 103:157–167.
Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR . Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 2005; 12:952–957.
Jones TA, Zou JY, Cowan SW, Kjeldgaard M . Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr 1991; A47:110–119.
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 1994; 50 (Pt 5):760–763.
Acknowledgements
We thank A Saxena at X12C at the National Synchrotron Light Source of the Brookhaven National Laboratory for assistance with data collection, and members of YAY′s group for technical support. SMW is an adjunct member of Temasek Life Sciences Laboratory. This work was supported by Temasek Life Sciences Laboratory and a research grant from the Singapore Ministry of Education (T208A3124 to YAY). YAY is a member of the Structural Biology and Proteomics Research Program supported by the National University of Singapore.
The atomic coordinates and structure factors (code 2ZKO) have been deposited in the Protein Data Bank (www.pdbj.org).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary information, Figure S1
Structural details of protein-RNA interactions. (PDF 1062 kb)
Rights and permissions
About this article
Cite this article
Cheng, A., Wong, S. & Yuan, Y. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res 19, 187–195 (2009). https://doi.org/10.1038/cr.2008.288
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2008.288
Keywords
This article is cited by
-
Viral silencing suppressors and cellular proteins partner with plant RRP6-like exoribonucleases
Virus Genes (2020)
-
Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition
Nature Communications (2018)
-
Non-structural protein 1 from avian influenza virus H9N2 is an efficient RNA silencing suppressor with characteristics that differ from those of Tomato bushy stunt virus p19
Virus Genes (2018)
-
Attenuation of the virulence of a recombinant influenza virus expressing the naturally truncated NS gene from an H3N8 equine influenza virus in mice
Veterinary Research (2016)
-
Biochemical and structural characterization of the interface mediating interaction between the influenza A virus non-structural protein-1 and a monoclonal antibody
Scientific Reports (2016)