Abstract
Background:
Colorectal cancer is a common malignancy and one of the leading causes of cancer-related deaths. The metabolism of omega fatty acids has been implicated in tumour growth and metastasis.
Methods:
This study has characterised the expression of omega fatty acid metabolising enzymes CYP4A11, CYP4F11, CYP4V2 and CYP4Z1 using monoclonal antibodies we have developed. Immunohistochemistry was performed on a tissue microarray containing 650 primary colorectal cancers, 285 lymph node metastasis and 50 normal colonic mucosa.
Results:
The differential expression of CYP4A11 and CYP4F11 showed a strong association with survival in both the whole patient cohort (hazard ratio (HR)=1.203, 95% CI=1.092–1.324, χ2=14.968, P=0.001) and in mismatch repair-proficient tumours (HR=1.276, 95% CI=1.095–1.488, χ2=9.988, P=0.007). Multivariate analysis revealed that the differential expression of CYP4A11 and CYP4F11 was independently prognostic in both the whole patient cohort (P=0.019) and in mismatch repair proficient tumours (P=0.046).
Conclusions:
A significant and independent association has been identified between overall survival and the differential expression of CYP4A11 and CYP4F11 in the whole patient cohort and in mismatch repair-proficient tumours.
Similar content being viewed by others
Main
Colorectal cancer is one of the major contributors to cancer-related mortality in the developed world (Siegel et al, 2014, 2016). The introduction of screening programmes and the development of new drugs have improved the survival rate of colorectal cancer patients; however, the average 5-year survival rate remains poor at 55% (Brenner et al, 2014). The characterisation of novel biomarker targets can further improve the survival rate, as it provides a better understanding of the complex molecular events underpinning tumour development and, if clinically validated, these biomarkers have potential roles in screening, diagnosis, prognosis and monitoring disease progression (Coghlin and Murray, 2015; Alnabulsi and Murray, 2016).
The CYP4 cytochrome P450 family of enzymes metabolises omega-3 and omega-6 fatty acids to biologically active eicosanoids that are implicated in tumour initiation, development and progression (Yu et al, 2011; Johnson et al, 2015). Arachidonic acid, an omega-6 fatty acid, is converted by CYP4A11 to 20-hydroxyicosatetraenoic acid (20-HETE), which is considered a key modulator in tumour progression, angiogenesis and metastasis (Ljubimov and Grant, 2005; Guo et al, 2007). CYP4F11 is not an efficient metaboliser of arachidonic acid compared with CYP4A11; however, it is the predominant CYP4 enzyme involved in the metabolism of omega 3-fatty acids (Dhar et al, 2008). The substrate specificity is not yet fully characterised for CYP4V2 and CYP4Z1 (Guengerich and Cheng, 2011). Despite the recognition of the involvement of omega fatty acids in tumourigenesis, the role of the cytochrome P450 enzymes involved in this pathway has received very limited attention in cancer biology (Panigrahy et al, 2010).
Using monoclonal antibodies we have developed the cytochrome P450 enzymes CYP4A11, CYP4F11, CYP4V2 and CYP4Z1; this study has profiled the expression of these enzymes by immunohistochemistry on a tissue microarray containing a large and well-characterised cohort of colorectal cancers. The expression profile of each enzyme was established by light microscopy using a semi-quantitative scoring system. The prognostic significance of each enzyme was determined by assessing the relationship between their expression in tumours and overall survival.
Materials and methods
Monoclonal antibody development
Monoclonal antibodies to CYP4A11, CYP4V2 and CYP4Z1 were developed using short synthetic peptides (Murray et al, 1998). Multiple sequence alignments of the amino acid sequences were performed on these enzymes and other CYP4 family members, to identify regions with the highest amino acid diversity. To avoid poorly antigenic sequences of amino acids (e.g., transmembrane region), a range of bioinformatics tools were used to predict and model hydrophilic, accessible and antigenic polypeptide sequences, as well as the secondary and tertiary structures of each enzyme (Supplementary Materials and Methods S1).
The amino acid sequences of peptides used to generate the antibodies and their location on each enzyme are specified in Supplementary Table S1. All peptides (Almac Sciences Ltd, Edinburgh, UK) were conjugated to ovalbumin for immunisations and to bovine serum albumin for the enzyme-linked immunosorbent assay (ELISA) screenings (Duncan et al, 1992). The immunisation via the subcutaneous route, the production of hybridomas and the ELISA screenings were carried out as previously described (Murray et al, 1996, 1998; Brown et al, 2014). The development of the monoclonal antibody to CYP4F11 has been described previously (Kumarakulasingham et al, 2005).
Immunoblotting
The specificity of the monoclonal antibodies was established by immunoblotting using whole-cell lysate (human embryonic kidney cells-HEK 293, Novus Biologicals, Cambridge, UK) overexpressing the relevant CYP as a positive control and lysates from cells containing empty vector as a negative control. Microsomal fractions prepared from human liver tissues (BD Gentest Human Liver Microsomes Pooled Male Donors 20 mg ml−1 catalogue number 452172, BD Biosciences, Bedford, MA, USA) were also used to carry out immunoblotting validation for each antibody. The immunoblotting was carried out as described, except that the polyvinylidene difluoride membrane was incubated overnight at 4 °C with undiluted monoclonal antibody (neat hybridoma tissue culture supernatant) and the secondary antibody, horseradish-peroxidase-conjugated anti-mouse IgG (Sigma-Aldrich, Dorset, UK), was diluted 1/3000 in phosphate-buffered saline–Tween-20 (Swan et al, 2016). When using liver microsomes, 30 μg of samples were loaded per lane compared with 5 μg when using overexpression lysate.
Patient cohort and colorectal cancer tissue microarray
The patient cohort was retrospectively acquired from the Grampian Biorepository (www.biorepository.nhsgrampian.org). The cohort is composed of tissue samples from 650 patients who had undergone surgery for primary colorectal cancers between 1994 and 2009, at Aberdeen Royal Infirmary (Aberdeen, UK), which is the principal teaching hospital of NHS Grampian. Patients who had received neoadjuvant chemotherapy and/or radiotherapy were excluded.
Survival time was defined to be the period in whole months from the date of surgery to the date of death from any cause (i.e., all-cause mortality). Survival data on a 6-monthly basis was updated from the NHS Grampian electronic patient management system and no patients were lost to follow-up. At the time (March 2012) of the censoring of patient outcome data, there had been 309 (47.5%) deaths and patients who were still alive were censored. The median survival was 103 months (95% CI=86–120 months), the mean survival was 115 months (95% CI=108–123 months) and the median follow-up time, calculated by the ‘reverse Kaplan–Meier’ method, was 88 months (95% CI=79–97 months). The clinico-pathological characteristics of the patients and their association with survival are described in Table 1.
Histopathology reporting was in accordance with The Royal College of Pathologists UK guidelines for the histopathological reporting of resection specimens of colorectal cancer, which includes guidance from version 5 of the tumour, node, metastasis staging system (Williams et al, 2007).
Blocks of formalin-fixed, paraffin-embedded tissue specimens were used to construct the tissue microarray as previously described (O’Dwyer et al, 2011; Brown et al, 2014; Swan et al, 2016). The histopathological processing of tissue specimens and the construction of the tissue microarray are described in Supplementary Materials and Methods S1
Immunohistochemistry
A Dako autostainer (Dako, Ely, UK) was used to perform the immunohistochemistry for each antibody using the Dako EnVision system (Dako; Kumarakulasingham et al, 2005, Brown et al, 2014). Antigen retrieval (microwaving in 10 mm citrate buffer pH 6.0 for 20 min) was performed for all antibodies, except CYP4A11. The immunohistochemistry procedure and the antigen retrieval are described in Supplementary Materials and Methods S1. A semi-quantitative scoring system was used to evaluate the intensity of immunostaining of each antibody (Kumarakulasingham et al, 2005; O’Dwyer et al, 2011; Brown et al, 2016; Swan et al, 2016). The scoring was conducted independently by two observers (RS and GIM) who were unaware of the clinical data and outcome. The assessment of cores was performed using light microscopy (Olympus BX 51, Olympus, Southend-on-Sea, Essex, UK). Simultaneous re-evaluation of the cores by both investigators was used to resolve any discrepancies in the scores (<5% of cases).
Assessment of MMR status
The status of mismatch repair protein (MMR) in the patient cohort was classified as either defective or proficient, based on the immunohistochemical assessment of MLH1 and MSH2 proteins (Brown et al, 2014).
Data analysis and statistics
The data were entered into an Excel 2013 spreadsheet before being analysed using IBM SPSS version 24 for Windows 7 (IBM, Portsmouth, UK). The following statistical tests were used: Mann–Whitney U-test, Wilcoxon signed-rank test, χ2-test, Kaplan–Meier survival analysis, log-rank test and Cox multivariate analysis (variables entered as categorical variables) including the calculation of hazard ratios (HRs) and 95% CIs. A probability value of P⩽0.05 was regarded as statistically significant. The survival analysis of the different patients groups was conducted using the log-rank test. The scores for each protein were dichotomised using the following cutoff points; negative staining vs positive staining, negative and weak staining vs moderate and strong staining, and strong staining vs negative/weak/moderate staining. Further details of data analysis and statistics are provided in Supplementary Materials and Methods S1.
Ethics
The use of colorectal tissue samples in this study was approved by the Grampian Biorepository scientific access group committee (Tissue request number 0002). No written consent was required from patients for the use of formalin fixed wax embedded tissue samples in the colorectal cancer tissue microarray.
Results
Monoclonal antibodies
During the hybridoma production, sequential ELISA screenings (immunogenic peptide specific to each enzyme) were used to determine the specificity of the monoclonal antibodies towards CYP4A11, CYP4V2 and CYP4Z1 (Duncan et al, 1992). Furthermore, immunoblotting showed a band migrating at the expected molecular weight for each antibody, whereas no band was detected in the negative controls (Supplementary Figure S1). The specificity of the antibody to CYP4F11 was confirmed previously (Kumarakulasingham et al, 2005).
Immunohistochemistry
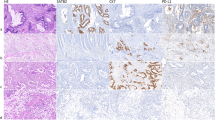
CYP4A11, CYP4F11 and CYP4V2 antibodies showed immunoreactivity in normal colonic epithelium, primary colorectal tumours and lymph node metastasis, whereas CYP4Z1 showed immunoreactivity only in a very small proportion of primary tumours. The immunostaining was exclusively localised to the cytoplasm of the cells (Supplementary Figure S2). Intra-tumour heterogeneity was not observed in either primary or metastatic colorectal tumours.
There was a significant increase in the intensity of immunostaining in primary tumours compared with normal colonic mucosa for CYP4A11 (P<0.001), CYP4F11 (P<0.001) and CYP4V2 (P<0.001) (Table 2 and Supplementary Figure S3). In contrast, a significant decrease in the expression of CYP4A11 (P=0.007), CYP4F11 (P<0.001) and CYP4V2 (P<0.001) was observed in lymph node metastasis compared with all primary tumours. There was also a significant decrease in the expression of CYP4A11 (P=0.002), CYP4F11 (P<0.001) and CYP4V2 (P<0.001) in lymph node metastasis compared with their corresponding primary Dukes C tumours.
Relationship with pathological parameters
The relationships between the main pathological parameters and the expression of CYP4A11, CYP4F11, CYP4V2 and CYP4Z1 are summarised in Supplementary Tables S2A–C. Both CYP4A11 (χ2=13.148, P=0.041) and CYP4V2 (χ2=24.474, P<0.001) showed significant associations with Dukes stage, but only CYP4V2 displayed a significant relationship with tumour stage (χ2=17.837, P=0.037). The expression of CYP4A11 was significantly associated with tumour site (χ2=15.703, P=0.015). CYP4F11 also showed significant associations with tumour site (χ2=20.947, P=0.002), tumour differentiation (χ2=8.5552, P=0.036) and MMR status (χ2=13.441, P=0.004).
Survival analysis
Whole patient cohort
Different cutoff points of the immunostaining scores were used to investigate the association between the expression of CYP4A11, CYP4F11 and CYP4V2, and overall survival (Supplementary Table S3). The expression of CYP4A11 showed a consistent and significant association with overall survival (Figure 1). Overall, increasing intensity of CYP4A11 immunostaining was significantly associated with poorer outcome (HR=1.135, 95% CI=1.032–1.249, χ2=9.080, P=0.028). When each level of the intensity groups of CYP4A11 expression was considered independently using one reference group (negative expression), strong CYP4A11 immunostaining was significantly associated with poorer outcome (HR=1.541, 95% CI=1.144–2.077, χ2=8.006, P=0.005) (Supplementary Table S4). The median survival was 137 months (95% CI undefined) and the mean was 132 months (95% CI=117–147 months) for CYP4A11-negative tumours (n=175), whereas the median survival was 75 months (95% CI=58–91 months) and the mean was 96 months (95% CI=84–109 months) for CYP4A11 strong expression tumours (n=197).
The overall relationship. The overall relationship between the expression of CYP4A11 and survival in the whole patient cohort using different cutoff points: negative vs weak vs moderate vs strong (A), further details of median survival times of individual groups, P-values and HRs are found in Supplementary Table S4), strong vs negative/weak/moderate (B), positive expression vs negative expression (C), and negative and weak vs moderate and strong (D).
Immunoreactivity for CYP4A11 was significantly associated with poorer prognosis (HR=1.346, 95% CI=1.032–1.756, χ2=4.881, P=0.027) when compared with CYP4A11-negative tumours. For CYP4A11-positive tumours (n=450), the median survival was 88 months (95% CI=71–104 months) and the mean was 105 months (95% CI=96–114 months), compared with a median of 137 (95% CI undefined) and a mean of 132 months (95% CI=117–147 months) for CYP4A11-negative tumours (n=175). Comparing strong CYP4A11-expressing tumours with negative/weak/moderate-expressing tumours also showed a significant association with survival (HR=1.379, 95% CI=1.089–1.746, χ2=7.234, P=0.007). The median survival was 113 months (95% CI=89–136 months) and the mean was 124 months (95% CI=114–134 months) for negative/weak/moderate CYP4A11-immunostaining tumours (n=428), whereas the median survival was 75 months (95% CI=58–91 months) and the mean was 96 months (95% CI=84–109 months) for strong CYP4A11-immunostaining tumours (n=197).
Exploratory analysis of CYP4 enzyme expression showed there was a significant association between the differential (combined) expression of CYP4A11 and CYP4F11, and survival (Supplementary Table S5). Therefore, a new variable, based on the differential expression of CYP4A11 and CYP4F11, was created to stratify tumours into three groups; CYP4A11>CYP4F11, CYP4A11=CYP4F11 and CYP4A11<CYP4F11. Overall survival was significantly associated with the expression profiles of CYP4A11>CYP4F11, CYP4A11=CYP4F11 and CYP4A11<CYP4F11 groups (HR=1.311, 95% CI=1.140–1.506, χ2=14.968, P=0.001) (Figure 2). When each level of the differential expression groups was considered independently using pairwise comparisons and one reference group (CYP4A11<CYP4F11), both CYP4A11>CYP4F11 (HR=1.733, 95% CI=1.306–2.300, χ2=14.405, P=<0.001) and CYP4A11=CYP4F11 (HR=1.432, 95% CI=1.064–1.928, χ2=5.425, P=0.020) were significantly associated with poorer outcome (Supplementary Table S6). The mean survival was 137 months (95% CI=124–151 months) (median survival undefined) for the CYP4A11<CYP4F11 group (n=214), the median survival was 95 months (95% CI=72–117 months) and the mean was 102 months (95% CI=90–114 months) for the CYP4A11=CYP4F11 group (n=185), whereas the median survival was 75 months (95% CI=60–89 months) and the mean survival was 94 months (95% CI=81–106 months) for CYP4A11>CYP4F11 group (n=217).
The overall relationship. The overall relationship between the differential expression of CYP4A11 and CYP4F11, and survival in the whole patient cohort (A), in MMR-proficient tumours (B) and in MMR-deficient tumours (C). Further details of median survival times of individual groups, P-values and HRs are found in Supplementary Table S6.
The associations between the expression of CYP4A11, CYP4F11, CYP4V2 and CYP4Z1, and overall survival in relation to different tumour sites, Dukes stage and extramural venous invasion are shown in Supplementary Tables S7–S10.
Mismatch repair protein-proficient cohort
There was a significant association between the expression of CYP4A11 and overall survival in MMR-proficient tumours (HR=1.156, 95% CI=1.040–1.286, χ2=11.221, P=0.011) (Figure 3 and Supplementary Table S11). When each level of the intensity groups of CYP4A11 expression was considered separately using pairwise comparisons and one reference group (negative expression), strong CYP4A11 immunoreactivity was significantly associated with poorer prognosis (HR=1.644, 95% CI=1.183–2.284, χ2=8.626, P=0.003) (Supplementary Table S4). When comparing strong CYP4A11-expressing tumours with negative/weak/moderate-expressing tumours, the strong expression of CYP4A11 showed a significant association with worse survival (HR=1.491, 95% CI=1.152–1.929, χ2=9.404, P=0.002). The positive expression of CYP4A11 was also significantly associated with poorer outcome when positive CYP4A11-expressing tumours were compared with negative CYP4A11-expressing tumours (HR=1.375, 95% CI=1.022–1.851, χ2=4.485, P=0.034).
The relationship between the expression of CYP4A11 and survival in MMR-proficient tumours. The relationship between the expression of CYP4A11 and survival in MMR-proficient tumours using different cutoff points: negative vs weak vs moderate vs strong (A), further details of median survival times of individual groups, P-values and HRs are found in Supplementary Table S4), strong vs negative/weak/moderate (B) and positive expression vs negative expression (C).
There was also a significant association between the differential expression of CYP4A11 and CYP4F11, and survival in MMR-proficient tumours (HR=1.276, 95% CI=1.05–1.488, χ2=9.988, P=0.007) (Figure 2). When each level of the intensity groups was considered independently using pairwise comparisons and one reference group (CYP4A11<CYP4F11), CYP4A11>CYP4F11-expressing tumours were significantly associated with poorer outcome (HR=1.629, 95% CI=1.199–2.214, χ2=9.261, P=0.002) (Supplementary Table S6). The median survival was 75 months (95% CI=85–121 months) and the mean was 97 months (95% CI=83–111 months) for CYP4A11>CYP4F11-expressing tumours (n=181), whereas the mean survival was 137 months (95% CI=123–151 months) (median survival undefined) for CYP4A11<CYP4F11-expressing tumours (n=186).
Mismatch repair protein-deficient cohort
The lack of expression of CYP4F11 was significantly associated with worse overall survival compared with CYP4F11-positive tumours (HR=0.479, 95% CI=0.241–0.952, χ2=4.682, P=0.03) (Supplementary Table S11 and Supplementary Figure S4). The median survival was 28 (95% CI=21–34 months) and the mean was 49 months (95% CI=28–70 months) for CYP4F11-negative tumours (n=16) compared with a median of 114 (95% CI=78–149 months) and a mean of 104 months (95% CI=84–123 months) for CYP4F11-positive tumours (n=77).
Overall, the association between survival and the differential expression of CYP4A11 and CYP4F11 just failed to reach the threshold for statistical significance in MMR-deficient cohort (HR=1.433, 95% CI=0.993–2.067, χ2=5.676, P=0.059) (Figure 2). When each level of the intensity groups was considered independently using pairwise comparisons and one reference group (CYP4A11<CYP4F11), both CYP4A11>CYP4F11-expressing tumours (HR=1.733, 95% CI=1.306–2.300, χ2=14.405, P=<0.001) and CYP4A11=CYP4F11-expressing tumours (HR=1.432, 95% CI=1.064–1.928, χ2=5.425, P=0.020) were significantly associated with poorer outcome (Supplementary Table S6).
Multivariate analysis
To evaluate the prognostic value of the differential expression of CYP4A11 and CYP4F11 (as a single variable) in relation to established prognostic parameters, multivariate analysis was performed using ‘Forward Stepwise: Conditional LR’ Cox regression method. The model showed there was a significant and independent prognostic value of using the differential expression of CYP4A11 and CYP4F11 in the whole patient cohort (P=0.019) and in MMR-proficient tumours (P=0.046) (Table 3 and Supplementary Tables S12 and S13). The differential expression was also a significant independent prognostic indicator in a multivariate analysis using only parameters that would be available at the time of biopsy in both the whole patient cohort (P=0.001) and in MMR-proficient tumours (P=0.006) (Supplementary Table S14).
Discussion
The rise in incidence and the poor survival rate makes colorectal cancer a major health burden in the developed world (Brenner et al, 2014; Siegel et al, 2014, 2016). There is still urgent need to identify and validate biomarkers of colorectal cancer that can play a role in clinical practice (Alnabulsi and Murray, 2016).
In this study, we have produced monoclonal antibodies to P450 enzymes CYP4A11, CYP4V2 and CYP4Z1 using short synthetic peptides that are specific to the targets of interest. The antibody for CYP4F11 was generated in a previous study (Kumarakulasingham et al, 2005). The antibodies were used to profile the expression of each enzyme by immunohistochemistry, which was performed on a well-characterised colorectal cancer tissue microarray.
The cytochrome P450 superfamily is classified into families, subfamilies and individual forms according to sequence homology and substrate specificity (Spector, 2009; Almira Correia et al, 2011; Fleming, 2011). Members of CYP1, CYP2 and CYP3 families are the major xenobiotic metabolising enzymes whose roles in cancer have been extensively studied (Murray et al, 1991, 1993, 1999, 2001, 2010; Rodriguez-Antona et al, 2010; Stenstedt et al, 2012; Xu et al, 2012). The CYP4 and higher numbered families are involved in the metabolism of a diverse range of endogenous compounds including eicosanoids, fatty acids, steroids and vitamins (Spector, 2009; Arnold et al, 2010; Panigrahy et al, 2010; Fleming, 2011; Guengerich and Cheng, 2011; Niwa et al, 2011). The role of CYP4 family and higher numbered families is not well studied in tumour biology with the exception of those CYPs involved in sex hormone metabolism in relation to breast and prostate cancer (Brueggemeier et al, 2005; Leroux, 2005; Stein et al, 2012). Therefore, this study aimed to examine the role of the main CYP4 family enzymes in colorectal cancer by characterising the expression of these enzymes using a large and well-characterised patient cohort.
This study revealed there was a significant increase in the expression of CYP4A11 in primary colorectal tumours compared with normal colonic mucosa and the increased expression was significantly associated with poorer prognosis. Consistent with our finding, an upregulation of CYP4A11 was demonstrated by a cDNA microarray-bioinformatics analysis of 10 colorectal tumours and their corresponding normal tissues (Yeh et al, 2006). Furthermore, the overexpression of CYP4A11 has been linked to rise in 20-HETE levels and upregulation of vascular endothelial growth factor (VEGF) and matrix metalloproteinases-9 (MMP-9) in non-small cell lung cancer (Yu et al, 2011). Both VEGF and MMP-9 are strong promoters of tumour invasion and metastasis (Yu et al, 2011; Goel and Mercurio, 2013; Brown and Murray, 2015). Previous research also showed that using selective inhibitors to downregulate the expression of CYP4A11 in cell lines and animal models resulted in a decrease in tumour growth, angiogenesis and metastasis of non-small cell lung cancer, renal adenocarcinoma and glioma (Guo et al, 2008, Alexanian et al, 2009, Yu et al, 2011). Our data have shown CYP4A11 is overexpressed in colorectal cancer; therefore, CYP4A11 may be a relevant therapeutic target in this type of cancer.
Comparing primary colorectal tumours to normal colonic mucosa also showed there was a significant increase in the expression of CYP4F11, which is a novel finding. In recent research, CYP4F11 expressed in cell lines (non-small cell lung cancer) converted oxalamides and benzothiazoles into stearoyl CoA desaturase (SCD) inhibitors (Theodoropoulos et al, 2016). Stearoyl CoA desaturase is emerging as a therapeutic target in cancer and therefore colorectal tumours with high CYP4F11 expression may be a valid target for SCD-targeted therapy.
The differential expression of CYP4A11 and CYP4F11 emerged as the best prognostic marker in this study. The distinct prognostic impact of the differential expression of CYP4A11 and CYP4F11 may be explained by differences in the enzymes substrates (Supplementary Figure S5). CYP4A11 converts arachidonic acid to metabolites that promote tumour growth and metastasis, whereas CYP4F11 metabolises omega 3-fatty acids to eicosanoids that inhibit tumour development and progression (Larsson et al, 2004; Kalsotra and Strobel, 2006; Dhar et al, 2008; Gelsomino et al, 2013; Barone et al, 2014). The differential expression of CYP4A11 and CYP4F11 was independently prognostic in multivariate analysis using the main prognostic parameters and also when only using information available at the time of biopsy diagnosis of colorectal cancer. Therefore, this biomarker combination could be a useful risk stratification tool especially if only tumour biopsies are available at the time of initial treatment decisions, which is a likely scenario considering colorectal cancer, especially rectal cancer, is moving towards neoadjuvant therapy followed by either observational follow-up or salvage surgery (Garcia-Aguilar et al, 2015).
The expression of each enzyme based on MMR status was also evaluated in this study, as this represents a major pathway in colorectal cancer (Boland and Goel, 2010; Geiersbach and Samowitz, 2011; Kim and Kim, 2014). Tumours lacking MMR proteins are already considered a distinct subgroup when dealing with prognosis and treatment of colorectal cancer (Hewish et al, 2010). Mismatch repair protein-proficient tumours represent the majority of colorectal cancer cases with a significantly worse prognosis than MMR-deficient tumours. Furthermore, novel promising treatments such as those targeting immune checkpoints have shown that MMR-proficient tumours are less responsive compared with MMR-deficient tumours (Le et al, 2015). Therefore, it is of particular interest to identify biomarkers for MMR-proficient tumours. In this study, the differential expression of CYP4A11 and CYP4F11 was significantly associated with prognosis in MMR-proficient tumours and, more importantly, both enzymes are actionable targets.
This study also found the expression of CYP4A11, CYP4F11 and CYP4V2 were significantly reduced in lymph node metastasis compared with their corresponding primary tumours. This provides further evidence to the concept that the phenotype of cancer cells is defined by their exposure to/and interaction with different microenvironment factors during their migration and within the metastatic site (Witz, 2008; Klein et al, 2012; Maman and Witz, 2013; Brown and Murray, 2015). The interrelationship between cancer cells and non-cancer cells within the microenvironment is increasingly acknowledged as a major factor in determining and understanding metastasis (McKay et al, 2000; Coghlin and Murray, 2010, 2014). The variation in the phenotypic expression between primary and metastatic tumours raises further doubts over the effectiveness of existing metastatic treatment models that is based on phenotypic assessment of primary tumour specimens.
In summary, CYP4A11, CYP4F11 and CYP4V2 are overexpressed in colorectal cancer, the increased expression of CYP4A11 is associated with poorer prognosis in both the total patient cohort and in MMR-proficient tumours, whereas the expression of CYP4F11 is associated with better outcome in MMR-deficient tumours. The differential expression of CYP4A11 and CYP4F11, which was independently prognostic in both the whole patient cohort and in MMR-proficient tumours, could provide the basis for a risk stratification tool in colorectal cancer. Furthermore, both enzymes are actionable drug targets and therefore could have therapeutic applications in colorectal cancer.
Change history
06 June 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Alexanian A, Rufanova VA, Miller B, Flasch A, Roman RJ, Sorokin A (2009) Down-regulation of 20-HETE synthesis and signaling inhibits renal adenocarcinoma cell proliferation and tumor growth. Anticancer Res 29: 3819–3824.
Almira Correia M, Sinclair PR, De Matteis F (2011) Cytochrome P450 regulation: the interplay between its heme and apoprotein moieties in synthesis, assembly, repair, and disposal. Drug Metab Rev 43: 1–26.
Alnabulsi A, Murray GI (2016) Integrative analysis of the colorectal cancer proteome: potential clinical impact. Expert Rev Proteomics 13: 917–927.
Arnold C, Konkel A, Fischer R, Schunck WH (2010) Cytochrome P450-dependent metabolism of omega-6 and omega-3 long-chain polyunsaturated fatty acids. Pharmacol Rep 62: 536–547.
Barone M, Notarnicola M, Caruso MG, Scavo MP, Viggiani MT, Tutino V, Polimeno L, Pesetti B, Di Leo A, Francavilla A (2014) Olive oil and omega-3 polyunsaturated fatty acids suppress intestinal polyp growth by modulating the apoptotic process in ApcMin/+ mice. Carcinogenesis 35: 1613–1619.
Boland CR, Goel A (2010) Microsatellite instability in colorectal cancer. Gastroenterology 138: 2073–2087.
Brenner H, Kloor M, Pox CP (2014) Colorectal cancer. Lancet 383: 1490–1502.
Brown GT, Cash B, Alnabulsi A, Samuel LM, Murray GI (2016) The expression and prognostic significance of bcl-2-associated transcription factor 1 in rectal cancer following neoadjuvant therapy. Histopathology 68: 556–566.
Brown GT, Cash BG, Blihoghe D, Johansson P, Alnabulsi A, Murray GI (2014) The expression and prognostic significance of retinoic acid metabolising enzymes in colorectal cancer. PLoS One 9: e90776.
Brown GT, Murray GI (2015) Current mechanistic insights into the roles of matrix metalloproteinases in tumour invasion and metastasis. J Pathol 237: 273–281.
Brueggemeier RW, Hackett JC, Diaz-Cruz ES (2005) Aromatase inhibitors in the treatment of breast cancer. Endocr Rev 26: 331–345.
Coghlin C, Murray GI (2010) Current and emerging concepts in tumour metastasis. J Pathol 222: 1–15.
Coghlin C, Murray GI (2014) The role of gene regulatory networks in promoting cancer progression and metastasis. Future Oncol 10: 735–748.
Coghlin C, Murray GI (2015) Biomarkers of colorectal cancer: recent advances and future challenges. Proteomics Clin Appl 9: 64–71.
Dhar M, Sepkovic DW, Hirani V, Magnusson RP, Lasker JM (2008) Omega oxidation of 3-hydroxy fatty acids by the human CYP4F gene subfamily enzyme CYP4F11. J Lipid Res 49: 612–624.
Duncan ME, McAleese SM, Booth NA, Melvin WT, Fothergill JE (1992) A simple enzyme-linked immunosorbent assay (ELISA) for the neuron-specific gamma isozyme of human enolase (NSE) using monoclonal antibodies raised against synthetic peptides corresponding to isozyme sequence differences. J Immunol Methods 151: 227–236.
Fleming I (2011) Cytochrome P450-dependent eicosanoid production and crosstalk. Curr Opin Lipidol 22: 403–409.
Garcia-Aguilar J, Renfro LA, Chow OS, Shi Q, Carrero XW, Lynn PB, Thomas CR Jr, Chan E, Cataldo PA, Marcet JE, Medich DS, Johnson CS, Oommen SC, Wolff BG, Pigazzi A, McNevin SM, Pons RK, Bleday R (2015) Organ preservation for clinical T2N0 distal rectal cancer using neoadjuvant chemoradiotherapy and local excision (ACOSOG Z6041): results of an open-label, single-arm, multi-institutional, phase 2 trial. Lancet Oncol 16: 1537–1546.
Geiersbach KB, Samowitz WS (2011) Microsatellite instability and colorectal cancer. Arch Pathol Lab Med 135: 1269–1277.
Gelsomino G, Corsetto PA, Campia I, Montorfano G, Kopecka J, Castella B, Gazzano E, Ghigo D, Rizzo AM, Riganti C (2013) Omega 3 fatty acids chemosensitize multidrug resistant colon cancer cells by down-regulating cholesterol synthesis and altering detergent resistant membranes composition. Mol Cancer 12: 137.
Goel HL, Mercurio AM (2013) VEGF targets the tumour cell. Nat Rev Cancer 13: 871–882.
Guengerich FP, Cheng Q (2011) Orphans in the human cytochrome P450 superfamily: approaches to discovering functions and relevance in pharmacology. Pharmacol Rev 63: 684–699.
Guo AM, Arbab AS, Falck JR, Chen P, Edwards PA, Roman RJ, Scicli AG (2007) Activation of vascular endothelial growth factor through reactive oxygen species mediates 20-hydroxyeicosatetraenoic acid-induced endothelial cell proliferation. J Pharmacol Exp Ther 321: 18–27.
Guo AM, Sheng J, Scicli GM, Arbab AS, Lehman NL, Edwards PA, Falck JR, Roman RJ, Scicli AG (2008) Expression of CYP4A1 in U251 human glioma cell induces hyperproliferative phenotype in vitro and rapidly growing tumors in vivo. J Pharmacol Exp Ther 327: 10–19.
Hewish M, Lord CJ, Martin SA, Cunningham D, Ashworth A (2010) Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat Rev Clin Oncol 7: 197–208.
Johnson AL, Edson KZ, Totah RA, Rettie AE (2015) Cytochrome P450 omega-hydroxylases in inflammation and cancer. Adv Pharmacol 74: 223–262.
Kalsotra A, Strobel HW (2006) Cytochrome P450 4F subfamily: at the crossroads of eicosanoid and drug metabolism. Pharmacol Ther 112: 589–611.
Kim ER, Kim Y (2014) Clinical application of genetics in management of colorectal cancer. Intest Res 12: 184–193.
Klein A, Sagi-Assif O, Izraely S, Meshel T, Pasmanik-Chor M, Nahmias C, Couraud P, Erez N, Hoon DS, Witz IP (2012) The metastatic microenvironment: Brain-derived soluble factors alter the malignant phenotype of cutaneous and brain-metastasizing melanoma cells. Int J Cancer 131: 2509–2518.
Kumarakulasingham M, Rooney PH, Dundas SR, Telfer C, Melvin WT, Curran S, Murray GI (2005) Cytochrome P450 profile of colorectal cancer: identification of markers of prognosis. Clin Cancer Res 11: 3758–3765.
Larsson SC, Kumlin M, Ingelman-Sundberg M, Wolk A (2004) Dietary long-chain n-3 fatty acids for the prevention of cancer: a review of potential mechanisms. Am J Clin Nutr 79: 935–945.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372: 2509–2520.
Leroux F (2005) Inhibition of p450 17 as a new strategy for the treatment of prostate cancer. Curr Med Chem 12: 1623–1629.
Ljubimov AV, Grant MB (2005) P450 in the angiogenesis affair: the unusual suspect. Am J Pathol 166: 341–344.
Maman S, Witz IP (2013) The metastatic microenvironment. In: Shurin MR, Umansky V, Malyguine A (eds). The Tumor Immunoenvironment. Springer Netherlands: Dordrecht, The Netherlands, 2013, pp 15–38.
McKay JA, Douglas JJ, Ross VG, Curran S, Ahmed FY, Loane JF, Murray GI, McLeod HL (2000) Expression of cell cycle control proteins in primary colorectal tumors does not always predict expression in lymph node metastases. Clin Cancer Res 6: 1113–1118.
Murray GI, Duncan ME, O’Neil P, Melvin WT, Fothergill JE (1996) Matrix metalloproteinase-1 is associated with poor prognosis in colorectal cancer. Nat Med 2: 461–462.
Murray GI, Duncan ME, Arbuckle E, Melvin WT, Fothergill JE (1998) Matrix metalloproteinases and their inhibitors in gastric cancer. Gut 43: 791–797.
Murray GI, Foster CO, Barnes TS, Weaver RJ, Ewen SW, Melvin WT, Burke MD (1991) Expression of cytochrome P450IA in breast cancer. Br J Cancer 63: 1021–1023.
Murray GI, McFadyen MC, Mitchell RT, Cheung YL, Kerr AC, Melvin WT (1999) Cytochrome P450 CYP3A in human renal cell cancer. Br J Cancer 79: 1836–1842.
Murray GI, Melvin WT, Greenlee WF, Burke MD (2001) Regulation, function, and tissue-specific expression of cytochrome P450 CYP1B1. Annu Rev Pharmacol Toxicol 41: 297–316.
Murray GI, Paterson PJ, Weaver RJ, Ewen SW, Melvin WT, Burke MD (1993) The expression of cytochrome P-450, epoxide hydrolase, and glutathione S-transferase in hepatocellular carcinoma. Cancer 71: 36–43.
Murray GI, Patimalla S, Stewart KN, Miller ID, Heys SD (2010) Profiling the expression of cytochrome P450 in breast cancer. Histopathology 57: 202–211.
Niwa T, Murayama N, Yamazaki H (2011) Stereoselectivity of human cytochrome p450 in metabolic and inhibitory activities. Curr Drug Metab 12: 549–569.
O’Dwyer D, Ralton LD, O’Shea A, Murray GI (2011) The proteomics of colorectal cancer: identification of a protein signature associated with prognosis. PLoS One 6: e27718.
Panigrahy D, Kaipainen A, Greene ER, Huang S (2010) Cytochrome P450-derived eicosanoids: the neglected pathway in cancer. Cancer Metastasis Rev 29: 723–735.
Rodriguez-Antona C, Gomez A, Karlgren M, Sim SC, Ingelman-Sundberg M (2010) Molecular genetics and epigenetics of the cytochrome P450 gene family and its relevance for cancer risk and treatment. Hum Genet 127: 1–17.
Siegel R, DeSantis C, Jemal A (2014) Colorectal cancer statistics, 2014. CA Cancer J Clin 64: 104–117.
Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66: 7–30.
Spector AA (2009) Arachidonic acid cytochrome P450 epoxygenase pathway. J Lipid Res 50 (Suppl): S52–S56.
Stein MN, Goodin S, Dipaola RS (2012) Abiraterone in prostate cancer: a new angle to an old problem. Clin Cancer Res 18: 1848–1854.
Stenstedt K, Hallstrom M, Johansson I, Ingelman-Sundberg M, Ragnhammar P, Edler D (2012) The expression of CYP2W1: a prognostic marker in colon cancer. Anticancer Res 32: 3869–3874.
Swan R, Alnabulsi A, Cash B, Alnabulsi A, Murray GI (2016) Characterisation of the oxysterol metabolising enzyme pathway in mismatch repair proficient and deficient colorectal cancer. Oncotarget 7: 46509–46527.
Theodoropoulos PC, Gonzales SS, Winterton SE, Rodriguez-Navas C, McKnight JS, Morlock LK, Hanson JM, Cross B, Owen AE, Duan Y, Moreno JR, Lemoff A, Mirzaei H, Posner BA, Williams NS, Ready JM, Nijhawan D (2016) Discovery of tumor-specific irreversible inhibitors of stearoyl CoA desaturase. Nat Chem Biol 12: 218–225.
Williams GT, Quirke P, Shepherd NA (2007) Dataset for Colorectal Cancer 2nd edn 1–27. The Royal College of Pathologists: London, UK.
Witz IP (2008) Tumor–microenvironment interactions: dangerous liaisons. Adv Cancer Res 100: 203–229.
Xu D, Hu J, De Bruyne E, Menu E, Schots R, Vanderkerken K, Van Valckenborgh E (2012) Dll1/Notch activation contributes to bortezomib resistance by upregulating CYP1A1 in multiple myeloma. Biochem Biophys Res Commun 428: 518–524.
Yeh C, Wang J, Cheng T, Juan C, Wu C, Lin S (2006) Fatty acid metabolism pathway play an important role in carcinogenesis of human colorectal cancers by microarray-bioinformatics analysis. Cancer Lett 233: 297–308.
Yu W, Chen L, Yang YQ, Falck JR, Guo AM, Li Y, Yang J (2011) Cytochrome P450 omega-hydroxylase promotes angiogenesis and metastasis by upregulation of VEGF and MMP-9 in non-small cell lung cancer. Cancer Chemother Pharmacol 68: 619–629.
Acknowledgements
The immunohistochemistry was performed with the support of the Grampian Biorepository (www.biorepository.nhsgrampian.org/). The antibodies were developed in collaboration with Vertebrate Antibodies Ltd (www.vertebrateantibodies.com) from whom they are now available commercially. RS was supported by the Jean Shanks Foundation. This study was supported by funding from SMART: Scotland award schemes of Scottish Enterprise.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
AA is a PhD student supported by Vertebrate Antibodies. BC and AA are employees of Vertebrate Antibodies. GM is a scientific advisor to Vertebrate Antibodies. RS declares no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Alnabulsi, A., Swan, R., Cash, B. et al. The differential expression of omega-3 and omega-6 fatty acid metabolising enzymes in colorectal cancer and its prognostic significance. Br J Cancer 116, 1612–1620 (2017). https://doi.org/10.1038/bjc.2017.135
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.135