Abstract
Background:
MicroRNAs (miRs) are involved in the regulation of many processes that contribute to malignancy, including cell proliferation, radiation resistance, invasion and metastasis. The role of miR-330-3p, an miR upregulated in breast cancer, remains unclear.
Methods:
We examine the association of miR-330-3p with distant relapse-free survival in the Oxford cohort of breast cancer patients. We also study miR-330-3p function using in vitro invasion and ex ovo metastasis assays. Using in vitro luciferase assays, we validate a novel target gene for miR-330-3p, Collagen And Calcium Binding EGF Domains 1 (CCBE1). We assess functional consequences of CCBE1 loss by using siRNA-mediated knockdown followed by in vitro invasion assays. Lastly, we examine the expression profile of CCBE1 in breast carcinomas in the Curtis and TCGA Breast Cancer data sets using Oncomine Platform as well as distant relapse-free and overall survival of patients in the Helsinki University breast cancer data set according to CCBE1 expression status.
Results:
miR-330-3p is enriched in breast cancer, and higher levels of miR-330-3p expression are associated with lower distant relapse-free survival in a cohort of breast cancer patients. Consistent with these observations, overexpression of miR-330-3p in breast cancer cell lines results in greater invasiveness in vitro, and miR-330-3p-overexpressing cells also metastasise more aggressively ex ovo. We identify CCBE1 as a direct target of miR-330-3p, and show that knockdown of CCBE1 results in a greater invasive capacity. Accordingly, in breast cancer patients CCBE1 is frequently downregulated, and its loss is associated with reduced distant relapse-free and overall survival.
Conclusions:
We show for the first time that miR-330-3p targets CCBE1 to promote invasion and metastasis. miR-330-3p and CCBE1 may represent promising biomarkers in breast cancer.
Similar content being viewed by others
Main
Breast cancer is the most common malignancy in women (excluding non-melanoma skin cancers), as well as the second most common cause of cancer deaths in women in the developed world (International Agency for Research on Cancer, W. GLOBOCAN, 2012). While patients present at various stages of the disease, metastases are responsible for the majority of the cancer deaths (Chambers et al, 2002; Weigelt et al, 2005). Biologically, generation of distant disease involves multiple complex mechanisms, such as epithelial to mesenchymal transition (EMT), invasion and migration, anoikis resistance, and angiogenesis (reviewed in Geiger and Peeper, 2009; Lamouille et al, 2014; Li and Li, 2015). Elucidation and understanding of such mechanisms is instrumental for the discovery of more accurate prognostic markers as well as novel therapies (Weigelt et al, 2005). Among the many molecular players described to date in metastatic disease, the roles of microRNAs (miRs) have begun to be appreciated more recently.
miRs are non-coding RNAs, which bind to the 3′ untranslated regions (UTRs) of their target mRNAs, and regulate the target protein expression by promoting degradation of mRNA transcripts or by inhibition of protein translation (Hammond, 2015). Pro-metastatic roles for miRs have been described in the processes of EMT, invasion, angiogenesis, anoikis resistance and intravasation (reviewed in Bouyssou et al, 2014; Takahashi et al, 2015). One such example is miR-10b which has been shown to target HoxD1 directly, resulting in downregulation of RhoC and greater invasiveness of breast cancer cells in vitro and metastasis in vivo (Ma et al, 2007). Other miRs known to promote metastasis or invasiveness include miR-24 (targeting PTPN9 and PTPRF) (Du et al, 2013), miR-93 (LATS2) (Fang et al, 2012), miR103/107 (DICER) (Martello et al, 2010), miR-125b (STARD13) (Tang et al, 2012), miR-373 (TXNIP 373) (Chen et al, 2015), miR-620 (HPGD) (Huang et al, 2013) and miR-95 (Huang et al, 2013) However, there remain a large number of miRs whose roles in cancer have yet to be discovered.
In this study, we investigate the pro-metastatic role of miR-330-3p in human breast cancer. Previous studies have suggested that miR-330-3p may regulate PI3K/Akt and Erk pathways by targeting SH3GL2 in glioblastoma cells, resulting in increased proliferation, apoptosis resistance and invasion (Qu et al, 2012; Yao et al, 2014). Another study suggested a similar pro-metastatic role for miR-330-3p in oesophageal cancer cell lines by targeting PDCD4 and conferring increased apoptosis resistance, invasiveness in vitro, as well as proliferative capacity in vivo and in vitro (Meng et al, 2015). Likewise, in non-small cell lung cancer cell lines, miR-330-3p overexpression resulted in an increase of cell proliferation by targeting EGR2 (Liu et al, 2015). However, any potential contribution of miR-330-3p in breast cancer remains unexplored. In this report, we demonstrate that miR-330-3p expression is increased in a human breast cancer cohort, and that its levels inversely correlate with distant disease-free survival. Consistent with this observation, miR-330-3p-overexpressing breast cancer cell lines invade more readily in vitro. miR-330-3p expression also promotes metastasis in a validated ex ovo model, the chick chorioallantoic membrane (CAM) assay (Leong et al, 2012).
In further exploration of the mechanism of miR-330-3p action, we show that Collagen And Calcium Binding EGF Domains 1 (CCBE1) is a direct target using a luciferase assay. Interestingly, CCBE1 was originally identified in a scanning copy number and gene expression study on the 18q21–qter chromosomal region of breast and prostate cancer cell lines (Yamamoto and Yamamoto, 2007). In this study, CCBE1 was among a list of genes whose expression was fully or partially lost in the cancer cell lines. Further studies established CCBE1 to be a secreted molecule involved in lymphangiogenesis during embryonal development (Hogan et al, 2009), and congenital CCBE1 mutations have been shown to cause Hennekam syndrome, a disorder characterised by lymphoedema, dysmorphic facies and mental retardation (Alders et al, 2009; Connell et al, 2010). However, few studies have explored the role of CCBE1 in malignancies, despite the original observation regarding CCBE1 loss in cancer cells. An additional study by Barton et al supports CCBE1 as a potential tumour-suppressor. The authors of the study recognised that CCBE1 is frequently inactivated by promoter hypermethylation in ovarian cancer (Barton et al, 2010). In their study, CCBE1 loss of expression was associated with an increase in in vitro cell migration, as well as decrease in relapse-free survival in patients with ovarian carcinoma, suggesting that CCBE1 may act as a tumour-suppressor. However, a more recent study described CCBE1 as a tumour-promoter in gastrointestinal stromal tumours (GISTs) (Tian et al, 2016), where higher levels of CCBE1 was correlated with higher risk degrees of GIST. Overexpression of CCBE1 was also associated with an enhancement of angiogenesis and poorer prognosis in patients. Nonetheless, the role of CCBE1 in breast cancer remains unclear. We provide evidence that CCBE1 may function a tumour-suppressor in breast cancer via in vitro invasion assays of human breast cancer cell lines. We also utilise breast cancer gene expression data sets to show examine CCBE1 expression in breast cancer patients, as well as correlation of CCBE1 with survival data. Taken together, the results of this study provide a novel role for miR-330-3p as a pro-metastatic microRNA, which targets CCBE1 to regulate metastasis in breast cancer. Our study provides new insight into the mechanisms by which breast cancer may develop a more aggressive phenotype.
Materials and methods
Cell lines and cell culture
Human breast adenocarcinoma (MDA-MB-231) cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Early passage cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g−1 glucose (Invitrogen, Burlington, ON, Canada) supplemented with 10% fetal bovine serum (Invitrogen) and penicillin (100 U ml−1)–streptomycin (100 μg ml−1) (Invitrogen) (hereafter referred to as 10% DMEM), and maintained in a humidified 37 °C incubator with 5% CO2. MCF10DCIS.COM cells (Miller et al, 2000) were obtained from Wayne State University, and cultured in DMEM/F12 supplemented with 1.05 mM calcium chloride (Invitrogen), 5% horse serum (Invitrogen), 10 mM HEPES (Invitrogen) and penicillin (100 U ml−1)–streptomycin (100 μg ml−1) (Invitrogen). Cell lines were passaged when they reached approximately 80% confluency and were regularly tested with MycoAlert (Lonza, Mississauga, ON, Canada) to ensure the absence of mycoplasma contamination.
Transfection of microRNA mimics and siRNA
3 × 105 cells were seeded into six-well plates, then 16 h later, miScript miR miR-330-3p mimic or miR control mimic (Thermo Fisher Scientific, Waltham, MA, USA) were mixed with 6 μl of DharmaFECT transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA) and DMEM as per the manufacturer’s instructions, then added to 10% DMEM for transfection of the cells. For siRNA transfections, control or a pool of three different siRNA for CCBE1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were transiently transfected into cells using Lipofectamine 2000 (Invitrogen) as per the manufacturer’s recommendations, and 24 h later, Matrigel invasion and migration assays (described below) were performed on the transfected cells.
Generation of stable overexpressing miR-330-3p cell lines
Cells were transduced with shMIMIC miR-330-3p or non-silencing control lentiviral particles as per the manufacturer’s instructions (Thermo Fisher Scientific, Philadelphia, PA, USA), selected using puromycin for 2 weeks and stable transductants were pooled; miR-330-3p overexpression was verified by real-time quantitative PCR.
Real-time quantitative PCR
For microRNA expression, total microRNA was extracted from cells or tumours using the mirVana miR kit (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesised using the miScript II RT kit (Qiagen, Toronto, ON, Canada) as per the manufacturer’s instructions. The mature miR-330-3p expression level was quantified through quantitative real-time PCR using the mi-Script SYBR Green PCR kit (Qiagen) and miScript Primer Assay for SNORD61 and miR-330-3p (Qiagen) on the StepOnePlus Real-time PCR system (Life Technologies, Burlington, ON, Canada). For gene expression, RNA was extracted using the RNeasy Mini kit (Qiagen) and cDNA synthesised using Omniscript RT kit (Qiagen) as per the manufacturer’s instructions. CCBE1 expression levels were quantified through quantitative real-time PCR using the QuantiTect SYBR Green PCR kit (Qiagen) on the StepOnePlus Real-time PCR system. For both microRNA and mRNA, expression levels were calculated using the comparative Ct method via StepOne Software (Life Technologies), and relative expression levels normalised to SNORD61 (for microRNA) or GAPDH (for mRNA)). Primer sequences were as follows: GAPDH-F: CAGCCTCAAGATCATCAGCA, GAPDH-R:GTCTTCTGGGTGGCAGTGAT; CCBE1-F:TACCGATATGACCGGGAGAG, CCBE1-R:AGCTGCCCAAGGTATTGATG. miR-330-3p and SNORD 61 primers were purchased from Qiagen (Hs_miR-330-3p_1 and Hs_SNORD_61_11 miScript Primer Assays; Qiagen).
Matrigel transwell invasion assay
Cells were serum starved overnight (0.1% DMEM), then 2 × 105 cells were seeded on top of 8 μm transwell inserts (BD Biosciences, Mississauga, ON, Canada) with 0.1% DMEM and pre-coated with 1 mg ml−1 Matrigel (Becton, Dickinson and Company, Mississauga, ON, Canada); 10% DMEM was used as a chemoattractant. After 24 h, cells that had invaded through the Matrigel-coated transwell inserts were fixed, stained by Kwik-Diff Stain (Thermo Fisher Scientific, Mississauga, ON, Canada) and number of invading cells counted under × 10 using a Leica DM LB2 microscope (Leica Microsystems, Richmond Hill, Ontario, Canada).
CAM assay
The chicken CAM assay for ex ovo metastasis and extravasation efficiency was performed as previously described (Kim et al, 2016). MDA-MB-231 cells stably expressing GFP and miR-330-3p or the control mimics were grown to ∼80–90% confluency. Both cell groups were resuspended in PBS at a concentration of 1.0 × 106 cells per ml. One hundred microlitres of each group’s cell suspension were injected intravenously into the CAM of day 13 embryos (n=3 per group) using a microinjector. Intravascular and extravasated cells were counted in a marked area by an aluminium foil window (1 inch × 1 inch) at T=0 and 24 h, respectively, using wide-field fluorescence microscopy and × 10 objective. At least 100 cells per region of interest were examined at T=0 h. Extravasation efficiency of each group per embryo was calculated by dividing the number of extravasated cells at T=24 h by the number of intravascular cells at T=0 h. Then, mean extravasation efficiencies in the embryos were calculated. Metastatic colonies in each embryo were also counted after an additional 7-day incubation post extravasation efficiency analysis (n=3 per group).
Alexa Fluor-594 gelatin coated coverslip production
About 50 μg ml−1 solution of poly-L-lysine (Sigma-Aldrich, Oakvillle, ON, Canada) was prepared and cooled on ice. Sterile 22 mm diameter glass coverslips were placed in a six-well plastic tissue culture plate, covered with 2 ml of 0.1 M NaOH and incubated for 5–10 min at room temperature. After washing 3 × with sterile PBS (pH 7.4), coverslips were coated with 1.5 ml of 50 μg ml−1 solution of poly-L-lysine and incubated for 20 min at room temperature. After aspirating poly-L-lysine solution, coverslips were washed 3 × with sterile PBS. Poly-L-lysine-coated coverslips were covered with 1.5 ml of 0.5% glutaraldehyde and incubated for 15 min at room temperature. Alexa Fluor-594-conjugated gelatin (Thermo Fisher Scientific, Mississauga, ON, Canada) was diluted in 0.2% unlabelled gelatin at a 10 : 1 ratio and placed at 37 °C in an incubator. Glutaraldehyde solution was aspired and coverslips were washed 3 × with sterile PBS. Eighty microlitres of the Diluted Alexa Fluor-594-conjugated gelatin mixture was placed onto each coverslip. Then coverslips were incubated for 10 min at room temperature in the dark. After incubation, coverslips were gently washed 2 × with sterile PBS and stored at 4 °C in the dark for up to 5 days.
In vitro actin stress fibre formation assay
Cells were seeded on Alexa Fluor-594 gelatin-coated coverslips at a density of 2 × 104 cells per ml and incubated for 4–24 h at 37 °C in an incubator. At the end of the incubation period, cell culture medium from each well was aspirated and coverslips were fixed with 4% paraformaldehyde at room temperature. Then coverslips were washed 3 × with sterile PBS. To label actin fibres in cells with Alexa Fluor 488-conjugated phalloidin, fixed cells were permeabilised with 1 ml of 0.1% Triton X-100 solution for 5 min at room temperature. Then, actin stress fibres were stained with 2 ml of Alexa Fluor 488-conjugated phalloidin solution (2 μl of Alexa Fluor 647-conjugated phalloidin solution was added to 2 ml of 5% skim milk PBS) for 1 h at room temperature. Coverslips were washed 3 × with sterile PBS and then mounted on slides to analyse actin stress fibre formation as per Martin et al (2012).
Gene array expression
Total RNA was isolated from MDA-MB-321 control and MDA-MB-321 stable cell lines using RNeasy Mini Kit (Qiagen) as per the manufacturer's instructions, and gene expression profiling performed by The Centre for Applied Genomics (The Hospital for Sick Children, Toronto, ON, Canada) using an Affymetrix GeneChip Human Gene 2.0 ST array (Thermo Fisher Scientific, Waltham, MA, USA). Gene expression array data were normalised using the default parameters in Affymetrix Expression Console Software (V.1.2, Thermo Fisher Scientific, Waltham, MA, USA).
Western blotting
Cells were lysed in ice-cold radioimmunoassay precipitation assay lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA pH 8.0, 0.5% (v/v) Triton X-100 and Complete protease inhibitor cocktail (Roche, Laval, QC, Canada)). Cell debris and insoluble material were removed by centrifugation at 12 000 g at 4 °C for 20 min. Following protein quantitation using the Bradford protein assay (Bio-Rad, Mississauga, ON, Canada), 25 μg of lysate was loaded per lane and proteins resolved by 4–20% gradient SDS–PAGE gel, wet-transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA), and the membranes were incubated in 5% nonfat dry milk in Tris-buffered saline Tween-20 (TBST) (10 mM Tris-Base, 150 mM NaCl, 0.05% Tween-20; pH 7.4) for 1 h at room temperature to block nonspecific antibody binding, followed by incubation with primary antibody in 5% milk in TBST overnight at 4 °C with gentle agitation. The membranes were washed three times for 10 min each in TBST, then incubated in TBST at room temperature for 1 h, followed by three 10-min washes with TBST. Protein-antibody binding on the membranes was detected with the use of enhanced chemiluminescence Plus solution (GE Healthcare Life Sciences, Montreal, QC, Canada) followed by exposure of the membranes to X-ray film (FujiFilm, Mississauga, ON, Canada). Antibodies against CCBE1 and β-actin were purchased from Sigma-Aldrich and Cell Signaling Technology (Danvers, MA, USA), respectively. Experiments were performed three independent times.
Luciferase assay
3′UTR luciferase assays were performed as described previously (Huang et al, 2015). Briefly, cells were transiently co-transfected with a CCBE1 3′UTR luciferase reporter plasmid or CCBE1 3′UTR luciferase reporter plasmid with mutations in the predicted miR-330-3p binding site, pcDNA3 vector constitutively expressing Renilla luciferase and miScript miR miR-330-3p mimic or control mimic. Twenty-four hours later, cells were processed for firefly luciferase and Renilla luciferase activity using the Dual Glo Luciferase Assay System (Promega, Madison, WI, USA). The firefly luciferase activity was normalised to the Renilla luciferase activity.
Statistical analysis
All statistical tests were two-sided, and the statistical analysis was performed using the GraphPad Prism version 5.0 program (GraphPad Software Inc., La Jolla, CA, USA). Statistical significance was defined as P<0.05, and NS=nonsignificant. The Student’s t-test was used to compare the mean values between two groups. Data are presented as mean values with standard deviations unless otherwise noted.
Results
miR-330-3p expression is increased in human breast cancer
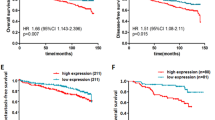
In order to investigate the association of miR-330-3p with breast cancer, we performed an miR expression analysis in a cohort of patients with early breast cancer (‘Oxford breast cohort’; GSE22216 (Buffa et al, 2011)). miR-330-3p levels were significantly elevated in breast cancer when compared with normal tissue (Figure 1A; log2: 2.01±0.18 (4.04-fold) relative expression in breast cancer versus healthy; **P<0.01). Moreover, 210 women from this cohort underwent 10-year follow-up, and we stratified the distant relapse-free survival data from these patients by levels of miR-330-3p. Analysis revealed significantly decreased rates of distant relapse in the lowest quartile of miR-330-3p expression versus the rest of the cohort (Figure 1B; log-rank test *P<0.05). Taken together, these results suggest that miR-330-3p expression is increased in breast cancer, and its expression may be a negative prognostic marker.
miR-330-3p is elevated in human breast cancer and is associated with reduced distant relapse-free survival. (A) Expression levels of miR-330-3p in a cohort of human breast cancer patients (GSE22216; cancer versus normal tissue). (B) The Kaplan–Meier curve of distant relapse-free survival in the early-stage breast cancer patient cohort: lowest quartile of miR-330-3p expression (solid) versus higher three quartiles (dashed) of miR-330-3p expression. A log-rank test was performed for statistical significance, and the hazard ratio was calculated between the two groups (*P<0.05, **P<0.01).
miR-330-3p expression increases cell invasiveness and metastatic potential
To study the influence of miR-330-3p on cellular invasion and metastasis formation directly, we first transiently transfected two human breast carcinoma cell lines (MDA-MB-321, MCF10DCIS.COM) with an miR-330-3p or a control mimic, and utilised the Matrigel transwell invasion assay (Huang et al, 2013, 2015; Mesci et al, 2014). In support of a pro-metastatic role for miR-330-3p, cell lines transfected with the miR-330-3p mimic exhibited significantly increased invasiveness in vitro in comparison with the control mimic (Figure 2A; MDA-MB-231: 2.4±0.3-fold increase over control, *P<0.05; MCF10DCIS.COM: 1.6±0.1-fold increase over control, *P<0.05). We then evaluated the influence of miR-330-3p on formation of metastases using a well-established ex ovo model, the chick CAM assay. This assay is designed to assess the propensity of cancer cell lines to migrate through the well-vascularised CAM and form micrometastases (Leong et al, 2012). For this purpose, we transduced the MDA-MB-231 cell line to overexpress miR-330-3p (MDA.330-3p) or the control (MDA.ctrl) mimic using a lentiviral system to ensure consistent levels of mimic expression. Similar to our observations earlier, miR-330-3p-expressing cells displayed a significantly greater ability to extravasate (Figure 2B; efficiency of 9.6±3.7% (miR-330-3p) versus 1.9±0.6% (control); *P<0.05), to form actin stress fibres (Figure 2B; 9.9±1.0 fibres per cell (miR-330-3p) versus 2.1±0.7 (control); ***P<0.001), and to generate micrometastatic colonies (Figure 2B; 8.7±1.3 colonies per embryo (miR-330-3p) versus 4.5±1.1 (control); *P<0.05). These data indicate that expression of miR-330-3p results in a more aggressive phenotype in human breast cancer by increasing cell invasiveness and formation of metastases, in vitro and ex ovo, respectively.
miR-330-3p increases invasiveness, extravasation and metastases. (A) MDA-MB-231 or MCF10.DCIS were transiently transfected with the miR-330-3p or the control mimic, followed by a Matrigel transwell invasion assay. Means, standard deviations and statistical significance are shown (*P<0.05; n=3 independent experiments). Representative images of the invasion assays for MDA-MB-231 are shown. Scale bar denotes 100 μm. (B) MDA-MB-231 cells were transduced with a lentivirus encoding for the control (MDA.ctrl) or the miR-330-3p (MDA.330p) mimic, followed by the chick CAM assay. Extravasation efficiency, actin stress fibres per cell and number metastatic colonies are shown with means, standard deviations and statistical significance (*P<0.05, ***P<0.001; n=3 independent experiments). Representative images are shown; green: MDA.ctrl or MDA.330p transductants (as labelled) expressing GFP; red: stroma stained with lectin; blue: stroma stained with dextran. Representative image shows three different planes of view (XY, XZ and YZ as labelled) Scale bar=20 μm, magnification: × 60.
miR-330-3p targets CCBE1 to promote invasiveness
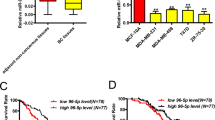
As miRs influence cellular behaviour by modulating the expression of target mRNAs, we generated a microarray expression profile from MDA-MB-321 cells transfected with the miR-330-3p or the control mimic to identify potential target genes. The gene array identified 974 putative targets for miR-330-3p. In parallel, we employed the TargetScan bioinformatics tool for miR target prediction (http://www.targetscan.org/vert_71/), which identified 4319 candidate target mRNAs for miR-330-3p. Then, we used Venny, a bioinformatics tool for comparing lists with Venn diagrams (http://bioinfogp.cnb.csic.es/tools/venny/; Oliveros, 2007–2015) to find genes that were common to both the gene array and TargetScan list, and found 37 possible candidates (Supplementary Figure S1). Interestingly, the previously published miR-330-3p targets, PDCD4, SH3GL2 and EGR2 were not identified by our method. We screened the 37 candidate mRNAs for a possible function as a tumour suppressor that could be targeted by miR-330-3p, and from this list, we selected CCBE1 for further validation. CCBE1 is a soluble extracellular matrix protein shown to play a role in lymph angiogenesis (Hogan et al, 2009); however, loss of CCBE1 is associated with greater invasiveness in ovarian cancer, as well as with reduced relapse-free survival (Barton et al, 2010). In agreement with the gene array and TargetScan results, miRWalk target prediction algorithm (Dweep et al, 2011, 2014) predicted that CCBE1 was a potential direct target of miR-330-3p (data not shown; 6 of 12 independent algorithms, including TargetScan). Comparison of miR-330-3p expression from the Oxford breast cohort (GSE22216) with CCBE1 transcript expression (GSE22219; mRNA data from Oxford breast cohort; Buffa et al, 2011) also revealed a statistically significant inverse correlation between miR-330-3p and CCBE1 (Figure 3A; Pearson’s rho=−0.129; ***P<0.005). Therefore, we proceeded to explore a potential regulatory axis involving miR-330-3p and CCBE1.
miR-330-3p targets CCBE1, and CCBE1 downregulation results in increased invasiveness. (A) Pearson correlation test of miR-330-3p expression with CCBE1 transcript expression from Oxford breast cohort (GSE22216 for miR and GSE22219 for mRNA); Pearson’s rho=−0.129, ***P<0.005. (B) MDA-MB-231 were transiently transfected with the miR-330-3p or the control mimic. Left panel: qPCR analysis of CCBE1 transcripts (normalised to GAPDH). Means and standard deviations are shown. Middle panel: representative western blot of CCBE1 protein levels (with β-actin control; numbers shown represent molecular weight markers in kDa). Right panel: densitometry analyses of CCBE1 protein normalised to β-actin control). (C) Sequence alignment of miR-330-3p with the wild type (wt) or mutated (mut) 3′UTR of CCBE1 showing the putative miR-330-3p binding site. MDA-MB-231 cells were transiently transfected with control or the miR-330-3p mimic, in combination with the wild type (wt; middle panel) or the mutated (mut; right panel) 3′UTR CCBE1 luciferase reporter. Luciferase activity was normalised to a renilla control. Means, standard deviations and statistical significance are shown. n=3 independent experiments. (D) MDA-MB-231 or MCF10DCIS.COM cells were transfected with a pool of CCBE1-specific siRNA (siCCBE1) or control (control) mimic, followed by qPCR analysis of CCBE1 transcripts (normalised to GAPDH) (left panel) or western blot analysis (normalised to β-actin) (middle panel – representative blot shown). Densitometry is shown in the right panel. (E) Matrigel transwell invasion assay of MDA-MB-231 or MCF10DCIS.COM cell lines transfected with siCCBE1 or Control as above. Means, standard deviations, and statistical significance are shown from n=3 independent experiments. Representative images are displayed (scale bar: MDA-MB-231: 200 μm; MCF10DCIS.COM: 500 μm).
In order to validate our findings biologically, we assessed CCBE1 levels in our MDA-MB-231 cells overexpressing miR-330-3p or the control mimic. CCBE1 transcripts were reduced with miR-330-3p overexpression (Figure 3B, left panel; 3.9±0.2-fold downregulation). In agreement, western blot analysis showed a decrease in CCBE1 protein levels with transfection of miR-330-3p mimic over control (Figure 3B, middle and right panels; 70% relative expression). Similarly, we confirmed reduced CCBE1 protein levels in the MDA.330-3p cells relative to MDA.ctrl (Supplementary Figure S2). Next, we proceeded with a luciferase reporter assay to confirm that miR-330-3p acts directly on the 3′UTR of CCBE1. To this end, we co-transfected a luciferase reporter vector expressing the wild type (wt) or mutated (mut) 3′UTR of CCBE1 into MDA-MB-231 cells together with miR-330-3p or control mimic (Figure 3C; sequence alignment of putative binding site of wild type or mutated 3′UTR of CCBE1 with miR-330-3p is shown). A reduction in normalised luciferase activity was observed in cells co-transfected with miR-330-3p relative to control (Figure 3C; 0.65±0.9-fold relative activity; **P<0.01). Conversely, when the predicted miR-330-3p binding site was mutated, no significant reduction of luciferase activity was noted (Figure 3C; 0.91±0.9-fold relative activity; NS). These results, therefore, establish CCBE1 as a direct target of miR-330-3p.
We then tested the effect of CCBE1 downmodulation directly by using a small interfering RNA (siRNA)-based strategy. We transfected MDA-MB-231 or MCF10DCIS.COM cells with a pool of three CCBE1-specific siRNA (‘siCCBE1’) or a control (‘control’) counterpart. CCBE1 transcripts were decreased in siCCBE1-transfected cells over control (Figure 3D; MDA-MB-231: 0.48±0.03-fold relative expression; MCF10DCIS.COM: 0.39±0.13-fold relative expression), in keeping with our earlier results. Downregulation on the transcript level was also confirmed by western blot analysis, which showed a reduction in CCBE1 protein (Figure 3D; middle and right panels; 71% relative expression). Functionally, siCCBE1-transfected cells showed greater invasiveness in vitro compared with their controls (Figure 3E; MDA-MB-231: 1.4±0.2-fold increase in invasion over control; *P<0.05; MCF10DCIS.COM: 3.07±0.7-fold increase in invasion over control; **P<0.01), as observed earlier with miR-330-3p-expressing cells. Collectively, these findings suggest that miR-330-3p directly targets CCBE1, ultimately resulting in increased invasiveness in human breast cancer cell lines.
CCBE1 is downregulated in breast carcinoma, and CCBE1 loss is associated with decreased survival
Next, we utilised Oncomine Platform (www.oncomine.com, September 2016), a repository of gene expression data sets of cancer patient samples, for possible correlations between CCBE1 expression and disease status. Comparison of normal breast tissue (n=144 patients) with ductal carcinoma in situ (DCIS; Figure 4A; 3.28-fold reduction; ***P<0.001. n=10 patients) and invasive ductal carcinoma (Figure 4A; 2.91-fold reduction; ***P<0.001. n=1556 patients) in the Curtis breast data set (Curtis et al, 2012) revealed lower levels of CCBE1 in the carcinoma samples. Similar to the Curtis data set, TCGA breast data set also revealed significantly lower levels of CCBE1 expression in invasive ductal carcinoma in comparison with normal breast (Figure 4A; 2.09-fold reduction; ***P<0.001; n=76 breast cancer and n=61 normal breast samples). To detect any associations of CCBE1 expression with survival in breast cancer patients, we also examined the data from a cohort of breast cancer patients from Helsinki University (GSE24450; Heikkinen et al, 2011; Muranen et al, 2011). Interestingly, patients whose tumours expressed lower levels of CCBE1 had poorer distant relapse-free and overall survival (Figure 4B left and middle panels; data stratified by lower 1 of 4 versus higher 3 of 4 of CCBE1 expression; **P<0.01, and *P<0.05, respectively. n=183 patients). In the Oxford breast cohort, there was a trend towards lower distant relapse-free survival in the low CCBE1 expression cohort, but this did not reach statistical significance (Figure 4B, right panel; data stratified by lower 1 of 4 versus higher 3 of 4 of CCBE1 expression; P=0.12 NS, n=216 patients). Collectively, these results support that CCBE1 loss may occur early in carcinogenesis, and that the extent of CCBE1 loss may represent a marker for poor prognosis in breast cancer in certain contexts.
CCBE1 is downregulated in a cohort of patients with DCIS and invasive ductal carcinoma, and CCBE1 levels inversely correlate with survival. (A) Oncotype Platform was searched for differential expression of CCBE1 in normal versus cancer tissues (gene keyword: CCBE1, search limited by breast cancer, cancer versus normal analysis). Results were ordered by fold change (reduction). Comparison of normal versus DCIS (left panel), normal versus invasive ductal carcinoma (middle panel) from Curtis Breast Cancer data set, as well as normal versus invasive ductal carcinoma from the TCGA Breast Cancer data set (right panel) are shown. 25–75 percentile range is shown by the box; median is indicated by the line inside the 25–75 percentile. Also shown are 10–90 percentile range (lines above and below the 25–75 percentile) and minimum/maximum values (dots). Fold change in CCBE1 expression and P-values is indicated. ***P<0.001. (B) Kaplan–Meier distant relapse-free survival (left panel) and overall survival (middle panel) curves of the Helsinki University breast cancer patient cohort (GSE24450) as well as distant relapse-free survival in the Oxford breast cohort (right panel; GSE22219) are shown as lower quartile of CCBE1 expression (dashed line) versus higher three quartiles of CCBE1 (solid line) expression. Log-rank test was performed for statistical significance (**P<0.01; *P<0.05; NS, nonsignificant, P>0.05).
Discussion
Numerous studies to date suggest that miRs are key players in metastatic disease, and thus understanding their mechanisms is of utmost importance (Ben-Hamo and Efroni, 2013; Bouyssou et al, 2014; Xue et al, 2014; Graveel et al, 2015; Takahashi et al, 2015). In this study, we demonstrate that miR-330-3p is an oncogenic molecule that shows a positive correlation with metastatic disease in breast cancer patient cohorts. miR-330-3p also increases the tendency of breast cancer cells to invade in vitro, as well as to form metastatic colonies. Mechanistically, we show that targeting of CCBE1 is a prominent mechanism for this phenotype. While CCBE1 has largely been studied in the context of Hennekam syndrome and lymphangiogenesis (Alders et al, 2009; Connell et al, 2010), few studies have examined its role in cancer (Barton et al, 2010; Tian et al, 2016). Biocomputational data suggest that CCBE1 may frequently be downregulated in breast cancers, although whether this loss of CCBE1 is related to miR-330-3p in patient samples is unclear at present. Nevertheless, lower levels of CCBE1 may prognosticate poorer survival, according to the Helsinki University breast cancer cohort. We report novel findings in the roles of miR-330-3p and CCBE1 in breast cancer metastasis, as well a new mode of regulation of CCBE1 expression by a miR, miR330-3p.
More generally, miR-330-3p is beginning to emerge as a tumour-promoting miR in a number of cancers. A small number of studies have shown that miR-330-3p can promote several hallmarks of cancer (e.g. proliferation, invasion) in glioblastoma, oesophageal carcinoma and non-small cell lung cancer (Qu et al, 2012; Yao et al, 2014; Liu et al, 2015; Meng et al, 2015). However, the role of miR-330-3p in breast cancer remained unexplored prior to our study. Furthermore, we include survival data from patient samples based on miR-330-3p expression, while previous studies exclusively relied upon in vitro, cell line-based data. In the context of the other cancers described above, a small number of miR-330-3p targets were identified, including SH3GL2 (Qu et al, 2012; Yao et al, 2014), PDCD4 (Meng et al, 2015) and EGR 2 (Liu et al, 2015). We provide evidence for a new connection between miR-330-3p and CCBE1. CCBE1-silenced cell lines behave similarly to miR-330-3p-overexpressing cell lines, and patient survival data suggest an inverse correlation between miR-330-3p and CCBE1 expression. These observations agree with the notion of CCBE1 as a target of miR-330-3p. Nonetheless, as miRs commonly have multiple targets and exert pleiotropic effects, we cannot exclude other possible targets that may have contributed to the phenotype we observed in our study. It is known that miRs can display varying effects in a tissue-dependent manner. For example, MDA-MB-231 cells do not express SH3GL2 transcripts, the target of miR-330-3p identified in glioblastoma (data not shown). Additionally, the levels of PDCD4 (identified in oesophageal carcinoma cell lines) do not change with miR-330-3p overexpression in MDA-MD-231 (data not shown), suggesting that in breast cancer, the target(s) of miR-330-3p are distinct from those previously identified. Another biological process that miR-330-3p could potentially target is EMT by modulating protein expression. However, we detected no significant difference in the relative abundance of the EMT markers Slug, Snail, Twist, Vimentin, Zeb-1 and Zeb-2 in MDA-MB-231 cells with miR-330-3p overexpression (data not shown). Future work may identify targets and cellular processes that provide a broader view of miR-330-3p as a tumour promoter.
While we provide the first report on CCBE1 loss in breast cancer, one notable study describing CCBE1 loss in ovarian cancers exists (Barton et al, 2010). This study showed a correlation with poorer survival with lower CCBE1 expression, similar to our results. CCBE1 loss occurred in all four FIGO stages of ovarian cancers, and the authors concluded that CCBE1 loss likely occurs early in carcinogenesis. Our results appear to be in concordance with this notion, as CCBE1 expression is significantly reduced in DCIS patient samples, a precursor lesion to invasive breast carcinoma. While CCBE1 downregulation in carcinomas and the associated phenotype agree between Barton et al and our study, the mechanisms of CCBE1 appears to differ. Our study shows a novel, miR-based regulation of CCBE1, while aberrant promoter hypermethylation was shown to be responsible for CCBE1 loss in the former study. While promoter status was not formally tested in our study, and a possible contribution by miR-330-3p was not explored by Barton et al, there may a variety of mechanisms by which CCBE1 expression is silenced in tumours. CCBE1 downregulation could thus represent a more general and consequential phenomenon in carcinogenesis than currently appreciated. In contrast, the only study available on the role of CCBE1 in GISTs (Tian et al, 2016) would suggest CCBE1 to be a tumour-promoter. In this study, higher levels of CCBE1 is associated with higher risk groups of GIST and poorer survival. The mechanistic basis for this appears to relate to tumour proliferation and angiogenesis, according to the authors’ findings. While this study appears to contradict our findings, as well as those by Barton et al (2010), the principal tumour histology and biology differs between this study and ours (ovarian and breast carcinomas versus mesenchymal tumour); CCBE1 action may thus be contextual based upon tumour origin. Consistent with this notion, in both our study and that by Barton et al, CCBE1 loss consistently confers greater cell migration/invasiveness, rather than altering proliferative capacity (data not shown). Future studies may clarify the specific biological contexts under which CCBE1 may act as a tumour-suppressor versus tumour promoter.
Ultimately, our understanding of the roles of miRs as well as some of their targets (such as CCBE1) in cancer are still in their early stages. Recognition of more miRs and their targets that contribute to aggressive cancer behaviour can lead to identification of cellular pathways that may aid in diagnostic, prognostic or personalised treatment approaches, and would be of a great value to scientist and clinician alike.
References
Alders M, Hogan BM, Gjini E, Salehi F, Al-Gazali L, Hennekam EA, Holmberg EE, Mannens MM, Mulder MF, Offerhaus GJ, Prescott TE, Schroor EJ, Verheij JB, Witte M, Zwijnenburg PJ, Vikkula M, Schulte-Merker S, Hennekam RC (2009) Mutations in CCBE1 cause generalized lymph vessel dysplasia in humans. Nat Genet 41 (12): 1272–1274.
Barton CA, Gloss BS, Qu W, Statham AL, Hacker NF, Sutherland RL, Clark SJ, O'Brien PM (2010) Collagen and calcium-binding EGF domains 1 is frequently inactivated in ovarian cancer by aberrant promoter hypermethylation and modulates cell migration and survival. Br J Cancer 102 (1): 87–96.
Ben-Hamo R, Efroni S (2013) MicroRNA-gene association as a prognostic biomarker in cancer exposes disease mechanisms. PLoS Comput Biol 9 (11): e1003351.
Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghobrial IM (2014) Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta 1845 (2): 255–265.
Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, Taylor M, Harris AL, Ragoussis J (2011) microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res 71 (17): 5635–5645.
Chambers AF, Groom AC, MacDonald IC (2002) Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2 (8): 563–572.
Chen D, Dang BL, Huang JZ, Chen M, Wu D, Xu ML, Li R, Yan GR (2015) MiR-373 drives the epithelial-to-mesenchymal transition and metastasis via the miR-373-TXNIP-HIF1alpha-TWIST signaling axis in breast cancer. Oncotarget 6 (32): 32701–32712.
Connell F, Kalidas K, Ostergaard P, Brice G, Homfray T, Roberts L, Bunyan DJ, Mitton S, Mansour S, Mortimer P, Jeffery S, Lymphoedema C (2010) Linkage and sequence analysis indicate that CCBE1 is mutated in recessively inherited generalised lymphatic dysplasia. Hum Genet 127 (2): 231–241.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Group M, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486 (7403): 346–352.
Du WW, Fang L, Li M, Yang X, Liang Y, Peng C, Qian W, O'Malley YQ, Askeland RW, Sugg SL, Qian J, Lin J, Jiang Z, Yee AJ, Sefton M, Deng Z, Shan SW, Wang CH, Yang BB (2013) MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J Cell Sci 126 (Pt 6): 1440–1453.
Dweep H, Gretz N, Sticht C (2014) miRWalk database for miRNA-target interactions. Methods Mol Biol 1182: 289–305.
Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk–database: prediction of possible miRNA binding sites by "walking” the genes of three genomes. J Biomed Inform 44 (5): 839–847.
Fang L, Du WW, Yang W, Rutnam ZJ, Peng C, Li H, O'Malley YQ, Askeland RW, Sugg S, Liu M, Mehta T, Deng Z, Yang BB (2012) MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle 11 (23): 4352–4365.
Geiger TR, Peeper DS (2009) Metastasis mechanisms. Biochim Biophys Acta 1796 (2): 293–308.
Graveel CR, Calderone HM, Westerhuis JJ, Winn ME, Sempere LF (2015) Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast Cancer (Dove Med Press) 7: 59–79.
Hammond SM (2015) An overview of microRNAs. Adv Drug Deliv Rev 87: 3–14.
Heikkinen T, Greco D, Pelttari LM, Tommiska J, Vahteristo P, Heikkila P, Blomqvist C, Aittomaki K, Nevanlinna H (2011) Variants on the promoter region of PTEN affect breast cancer progression and patient survival. Breast Cancer Res 13 (6): R130.
Hogan BM, Bos FL, Bussmann J, Witte M, Chi NC, Duckers HJ, Schulte-Merker S (2009) Ccbe1 is required for embryonic lymphangiogenesis and venous sprouting. Nat Genet 41 (4): 396–398.
Huang X, Taeb S, Jahangiri S, Emmenegger U, Tran E, Bruce J, Mesci A, Korpela E, Vesprini D, Wong CS, Bristow RG, Liu FF, Liu SK (2013) miRNA-95 mediates radioresistance in tumors by targeting the sphingolipid phosphatase SGPP1. Cancer Res 73 (23): 6972–6986.
Huang X, Taeb S, Jahangiri S, Korpela E, Cadonic I, Yu N, Krylov SN, Fokas E, Boutros PC, Liu SK (2015) miR-620 promotes tumor radioresistance by targeting 15-hydroxyprostaglandin dehydrogenase (HPGD). Oncotarget 6 (26): 22439–22451.
International Agency for Research on Cancer, W. GLOBOCAN (2012) Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012 http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx?cancer=breast (accessed 10 July 2015).
Kim Y, Williams KC, Gavin CT, Jardine E, Chambers AF, Leong HS (2016) Quantification of cancer cell extravasation in vivo. Nat Protoc 11: 937–948.
Lamouille S, Xu J, Derynck R (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15 (3): 178–196.
Leong HS, Chambers AF, Lewis JD (2012) Assessing cancer cell migration and metastatic growth in vivo in the chick embryo using fluorescence intravital imaging. Methods Mol Biol 872: 1–14.
Li L, Li W (2015) Epithelial-mesenchymal transition in human cancer: comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol Ther 150: 33–46.
Liu X, Shi H, Liu B, Li J, Liu Y, Yu B (2015) miR-330-3p controls cell proliferation by targeting early growth response 2 in non-small-cell lung cancer. Acta Biochim Biophys Sin (Shanghai) 47 (6): 431–440.
Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449 (7163): 682–688.
Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, Parenti AR, Daidone MG, Bicciato S, Piccolo S (2010) A MicroRNA targeting dicer for metastasis control. Cell 141 (7): 1195–1207.
Martin KH, Hayes KE, Walk EL, Ammer AG, Markwell SM, Weed SA (2012) Quantitative measurement of invadopodia-mediated extracellular matrix proteolysis in single and multicellular contexts. J Vis Exp (66): e4119.
Meng H, Wang K, Chen X, Guan X, Hu L, Xiong G, Li J, Bai Y (2015) MicroRNA-330-3p functions as an oncogene in human esophageal cancer by targeting programmed cell death 4. Am J Cancer Res 5 (3): 1062–1075.
Mesci A, Taeb S, Huang X, Jairath R, Sivaloganathan D, Liu SK (2014) Pea3 expression promotes the invasive and metastatic potential of colorectal carcinoma. World J Gastroenterol 20 (46): 17376–17387.
Miller FR, Santner SJ, Tait L, Dawson PJ (2000) MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst 92 (14): 1185–1186.
Muranen TA, Greco D, Fagerholm R, Kilpivaara O, Kampjarvi K, Aittomaki K, Blomqvist C, Heikkila P, Borg A, Nevanlinna H (2011) Breast tumors from CHEK2 1100delC-mutation carriers: genomic landscape and clinical implications. Breast Cancer Res 13 (5): R90.
Oliveros JC (2007–2015) "Venny”. An interactive tool for comparing lists with Venn's diagrams http://bioinfogp.cnb.csic.es/tools/venny/index.html.
Qu S, Yao Y, Shang C, Xue Y, Ma J, Li Z, Liu Y (2012) MicroRNA-330 is an oncogenic factor in glioblastoma cells by regulating SH3GL2 gene. PLoS ONE 7 (9): e46010.
Takahashi RU, Miyazaki H, Ochiya T (2015) The roles of microRNAs in breast cancer. Cancers (Basel) 7 (2): 598–616.
Tang F, Zhang R, He Y, Zou M, Guo L, Xi T (2012) MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7 and MDA-MB-231 breast cancer cells. PLoS ONE 7 (5): e35435.
Tian GA, Zhu CC, Zhang XX, Zhu L, Yang XM, Jiang SH, Li RK, Tu L, Wang Y, Zhuang C, He P, Li Q, Cao XY, Cao H, Zhang ZG (2016) CCBE1 promotes GIST development through enhancing angiogenesis and mediating resistance to imatinib. Sci Rep 6: 31071.
Weigelt B, Peterse JL, van 't Veer LJ (2005) Breast cancer metastasis: markers and models. Nat Rev Cancer 5 (8): 591–602.
Xue J, Niu J, Wu J, Wu ZH (2014) MicroRNAs in cancer therapeutic response: friend and foe. World J Clin Oncol 5 (4): 730–743.
Yamamoto F, Yamamoto M (2007) Scanning copy number and gene expression on the 18q21-qter chromosomal region by the systematic multiplex PCR and reverse transcription-PCR methods. Electrophoresis 28 (12): 1882–1895.
Yao Y, Xue Y, Ma J, Shang C, Wang P, Liu L, Liu W, Li Z, Qu S, Li Z, Liu Y (2014) MiR-330-mediated regulation of SH3GL2 expression enhances malignant behaviors of glioblastoma stem cells by activating ERK and PI3K/AKT signaling pathways. PLoS One 9 (4): e95060.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-Non-Commercial-Share Alike 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Mesci, A., Huang, X., Taeb, S. et al. Targeting of CCBE1 by miR-330-3p in human breast cancer promotes metastasis. Br J Cancer 116, 1350–1357 (2017). https://doi.org/10.1038/bjc.2017.105
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.105
Keywords
This article is cited by
-
Identifying the driver miRNAs with somatic copy number alterations driving dysregulated ceRNA networks in cancers
Biology Direct (2023)
-
Extracellular matrix remodeling in tumor progression and immune escape: from mechanisms to treatments
Molecular Cancer (2023)
-
Cancer-associated fibroblasts facilitate breast cancer progression through exosomal circTBPL1-mediated intercellular communication
Cell Death & Disease (2023)
-
CircSEMA4B inhibits the progression of breast cancer by encoding a novel protein SEMA4B-211aa and regulating AKT phosphorylation
Cell Death & Disease (2022)
-
Gene networks and transcriptional regulators associated with liver cancer development and progression
BMC Medical Genomics (2021)