Abstract
The current therapy success of depressive disorders remains in need of improvement due to low response rates and a delay in symptomatic improvement. Reliable functional biomarkers would be necessary to predict the individual treatment outcome. On the basis of the neurotrophic hypothesis of antidepressant’s action, effects of antidepressant drugs on proliferation may serve as tentative individual markers for treatment efficacy. We studied individual differences in antidepressant drug effects on cell proliferation and gene expression in lymphoblastoid cell lines (LCLs) derived from patients treated for depression with documented clinical treatment outcome. Cell proliferation was characterized by EdU (5-ethynyl-2'-deoxyuridine) incorporation assays following a 3-week incubation with therapeutic concentrations of fluoxetine. Genome-wide expression profiling was conducted by microarrays, and candidate genes such as betacellulin—a gene involved in neuronal stem cell regeneration—were validated by quantitative real-time PCR. Ex vivo assessment of proliferation revealed large differences in fluoxetine-induced proliferation inhibition between donor LCLs, but no association with clinical response was observed. Genome-wide expression analyses followed by pathway and gene ontology analyses identified genes with different expression before vs after 21-day incubation with fluoxetine. Significant correlations between proliferation and gene expression of WNT2B, FZD7, TCF7L2, SULT4A1 and ABCB1 (all involved in neurogenesis or brain protection) were also found. Basal gene expression of SULT4A1 (P=0.029), and gene expression fold changes of WNT2B by ex vivo fluoxetine (P=0.025) correlated with clinical response and clinical remission, respectively. Thus, we identified potential gene expression biomarkers eventually being useful as baseline predictors or as longitudinal targets in antidepressant therapy.
Similar content being viewed by others
Introduction
The therapy of depression is characterized by response rates around 60% and difficulties in the early evaluation of individual therapy success owing to delayed clinical improvement that may take from weeks up to several months.1, 2 So far, it is not yet possible to predict the individual treatment outcome of depressive patients owing to a lack of predictive biomarkers. According to the neuroplasticity hypothesis of the antidepressants’ action, which is based on both animal and human cell models, antidepressants act—at least in part—by increasing proliferation of neuronal stem cells.3, 4 Furthermore, depressed patients have been reported with volume reductions in hippocampus and other brain regions,5 which has been observed to be reversed after successful antidepressant therapy, apparently owing to antidepressant-induced triggering of neural plasticity.6 As cerebral remodeling processes are complex and take many weeks, this explains the observed delay in symptomatic improvement.7 Consequently, the late-onset action of antidepressant drugs in the treatment of depression is hypothesized owing to changes in neuroplasticity resulting from these proliferative effects in the hippocampus.8
Individual differences in antidepressant-mediated modulation of cell growth were observed in human blood-derived lymphoblastoid cell lines (LCLs): sensitivity to paroxetine was measured by effects on ex vivo cell proliferation to identify potential gene and miRNA antidepressant response biomarkers.9, 10 Assuming that inter-individual variations in antidepressant effects on cell proliferation rates may serve as surrogate indicators for individual treatment efficacy,8, 11, 12 we used LCLs from depressed patients to study the effect of fluoxetine on cellular proliferation rates and their association with clinical response data. Furthermore, genome-wide gene expression analyses may further be used to identify tentative cell proliferation-associated biomarkers. Here we applied phenotypic screening of antidepressant effects on cell proliferation, combined with genome-wide expression profiling, for identifying tentative antidepressant response biomarkers that may assist in the early identification of treatment-resistant depression patients.
Materials and methods
Patients and cell lines
Epstein–Barr virus-transformed LCLs were generated in a subset of patients from the Munich Antidepressant Response Signature (MARS) project. The MARS study is a naturalistic clinical study on antidepressant drug response designed for pharmacogenetics analyses of antidepressant drug response biomarkers as described earlier.2, 13, 14 From the available LCLs from patients, various cell lines were picked for experiments (n=10 for microarray analysis, and n=25 responder and n=25 non-responder to antidepressant drug treatment for proliferation phenotyping). For cell line selection, the response and non-response statuses were considered after 8 weeks of antidepressant drug treatment (study population parameters are summarized in Table 1; drug response profiles are listed in Supplementary Figure 1 and Supplementary Table 2). LCLs were gained by Epstein–Barr virus transformation from full EDTA (ethylenediaminetetraacetic acid)-blood samples provided by the MARS patients admitted to the hospital of the Max Planck Institute of Psychiatry in Munich, Germany, for depression treatment.2 The study was approved by the Ethical Committee of the Medical Faculty at the Ludwig-Maximilian University. The participating patients gave verbal and written informed consent to provide biomaterial for the study of antidepressant response biomarkers also including transformation of blood lymphocytes into cell lines. MARS is an observational study of depressed patients being treated according to the attending physician’s choice. Depressive symptoms were rated by the 21-item HDRS (Hamilton Depression Rating Scale) at weeks 0, 5 and 8 after study inclusion.15 Response was defined as HDRS reduction of at least 50% (compared with initial values at study inclusion) and remission was defined as a total reduction of HDRS to values smaller than 8.16
Generation and cultivation of lymphoblastoid cell lines
LCLs were generated from lymphocytes isolated from blood samples through Epstein–Barr virus transformation.17, 18 Peripheral blood mononuclear cells were isolated by density gradient centrifugation using Ficoll, resuspended in Epstein–Barr virus supernatant from B95-8 cell line, and 100 μl were seeded into wells of a 48-well cell culture plate. After the addition of 200 μl Roswell Park Memorial Institute (RPMI) medium (containing 20% fetal calf serum (FCS)) per well, the cells were incubated at 37 °C in a humidified CO2 incubator (with 5% CO2) for 5 days. Subsequently, one volume of fresh RPMI medium (containing 20% FCS) and cyclosporine A (Sigma-Aldrich, St Louis, MO, USA) in a final concentration of 1 μg ml−1 were added. On day 25 after isolation, the cells from different wells of the same sample were pooled and further cultivated with exchange of the medium (containing 15% FCS) every second day. The cell identity was tested using the T- and B-cell specific antibodies CD3, CD19 and CD45 (BD Tritest Kit, Becton Dickinson, Heidelberg, Germany) through flow cytometry. The cells were cryo-stored in 90% FCS and 10% dimethyl sulfoxide. The LCLs were cultured in RPMI medium supplemented with 15% FCS, antibiotics (100 μg ml−1 penicillin, 100 μg ml−1 streptomycin) and a final concentration of 4 mm l-glutamine. The cells were incubated at 37 °C in a humidified CO2 incubator (with 5% CO2) in cell culture flasks.
Incubation with antidepressants and LCL cell proliferation assay
EdU incorporation assays (Life Technologies, Carlsbad, CA, USA) were carried out according to the manufacturer instructions in technical and biological duplicates. Fluoxetine was chosen as antidepressant drug because its proliferative features are well studied,19, 20, 21 it showed the most distinct effects in preceding experiments using LCLs, and most of the MARS patients under antidepressant monotherapy received selective serotonin reuptake inhibitor antidepressant drugs. Mock-treated control cultures were grown in parallel, and cell density was set to 3 × 105 cells per milliliter every second day. The incubation periods and fluoxetine effects on cell proliferation were tested at 7, 14 and 21 days of incubation, and it turned out that largest effects were observed after 21 days of continuous incubation with fluoxetine at a concentration of 0.5 μg ml−1 (including change of fresh medium every second day). The period of 21 days incubation with fluoxetine was therefore chosen for ex vivo phenotyping of the entire LCL panel. Fluoxetine was purchased from Sigma-Aldrich and stock solutions were prepared in dimethyl sulfoxide.
Nucleic acid extraction
Nucleic acid extraction was performed with the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany). Nucleic acid concentrations were quantified using a NanoDrop Spectrophotometer (Thermo Scientific, Darmstadt, Germany).
Whole genome expression profiling
Microarray analyses were performed in 10 cell lines (untreated, and after 21 days of incubation with 0.5 μg ml−1 fluoxetine) using Agilent One Color Microarray Technology (Waldbronn, Germany; SurePrint G3 Human Gene Expression 8 × 60K Microarray Kit) containing probes for >27 000 transcripts. RNA quality was determined with Agilent 2100 Bioanalyzer and a total of 100 ng RNA was used for reverse transcription and labeling. The generation of complementary DNA (cDNA) was conducted with T7 promoter primers in a total reaction volume of 10 μl (containing 0.1 μm DTT, 5 μm dNTP mix and 1.2 μl RNase inhibitor in first-strand buffer) incubated for 2 h at 40 °C followed by 15 min at 70 °C. The labeling was performed for 2 h at 40 °C after the addition of NTP mix, T7 RNA polymerase and cyanin 3-CTP. After column-based purification of labeled complementary RNA, hybridization was carried out for 17 h and fluorescence intensities were measured by SureScan Microarray Scanner (Agilent). Data were normalized and summarized with the multiaverage method. Data analysis was conducted using GeneSpring (Agilent) and initially, the probeset was filtered on data files (control type 0) with the condition that at least 100% of the values in any one condition are within the expected range.
The differential gene expression was rated in pairs with fold-change cutoff of 2 and significance value of P<0.05 (uncorrected). Pathway analysis (single-experiment analysis) was performed using the imported pathway database from GenMAPP Pathway Markup Language and an uncorrected P-value cutoff of P<0.05 and focused on pathways inversely regulated in responder and non-responder indicator cell lines. Indicator cell lines are characterized by a rectified clinical response status, proliferation status and hierarchical clustering status: cell lines 24DC and 275U served as positive indicator cell lines and derived from clinical responders, they were in vitro proliferators and showed strong gene expression changes after treatment with fluoxetine, whereas cell line 278H was used as a negative indicator cell line. Gene ontology analysis was carried out using the web-based STRING database.22 Systematic search of central nervous system annotations were carried out using the gene names and one of the following terms: brain, neuron, neurogenesis, neural plasticity, proliferation, depression or antidepressant. Microarray data were deposited in NCBI’s Gene Expression Omnibus database23, 24 and are accessible through GEO Series accession number GSE83386 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE83386).
Gene expression analysis of candidate genes identified from genome-wide expression analyses
After cDNA preparation with 1 μg RNA using Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany) in a Gradient Mastercycler (Eppendorf, Hamburg, Germany) thermocycler (10 min at 25 °C, 30 min at 55 °C, 5 min at 85 °C), gene expression was measured through real-time PCR with the QuantiTect SYBR Green PCR kit (Qiagen) in a Light Cycler 480 real-time PCR instrument (95 °C for 10 min, followed by 60 cycles of 95 °C for 10 s, and 55 °C for 15 s, 72 °C for 20 s) in technical and biological duplicates. QuantiTect Primers were purchased from Qiagen, custom-made primers from Eurofins Genomics (Ebersberg, Germany; see Supplementary Table 1). The gene expression fold-change values were calculated by the ΔΔCT method using GAPDH as reference gene,25 whereas basal gene expression was indicated as ΔCT values of untreated samples.
Statistical analyses
Associations between LCL proliferation rates vs donor age and proliferation vs donor gender were calculated using Pearson’s correlation and Student’s t-test for equal variances (confirmed by Levene’s test), respectively. Unpaired Student’s t-test values were used to analyze the significance of basal gene expression differences between non-proliferating and proliferating cell lines in the edge-group approach, and to analyze basal gene expression differences between clinical subgroups in all the cell lines (clinical response after 5 and 8 weeks, and remission after 5 and 8 weeks). Data of gene expression fold-change values were analyzed by the Wilcoxon–Mann–Whitney rank-sum test. Statistical power amounts to 93.4% for EdU phenotyping experiments, to 63.57% for microarray experiments and to 99.9% for RT-PCR (PCR with reverse transcription) validation experiments with effect sizes of r=2 and significant levels of α=0.05 each. Statistical tests were calculated as two-sided and error bars are shown as standard deviations. For all the remaining applications, implemented statistics programs of the specific software (GeneSpring, STRING) were used. In general, P-values <0.05 were considered as significant and are reported as unadjusted unless stated otherwise. All Statistical analyses were carried out using IBM SPSS Statistics 21 (Ehningen, Germany).
Results
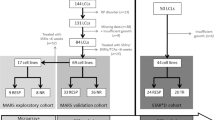
The experimental design consisted of an exploration and a validation phase (Figure 1). In the explorative phase genome-wide gene expression profiling and EdU proliferation phenotyping experiments were carried out in to identify potential gene expression biomarkers and to elucidate a possible association between individual antidepressant-induced LCL proliferation and clinical response from LCL donors, respectively. During the validation phase, both approaches were combined in an edge-group approach where the identified tentative gene expression biomarkers were evaluated in extreme cell lines from EdU phenotyping. Subsequently, the gene expression differences of the remaining candidate genes were determined between non-responder- and responder-derived cell lines.
Genome-wide gene expression profiling
The gene expression changes following 21-day fluoxetine treatment of n=10 LCLs (derived from n=6 responders and n=4 non-responders representing the average patient population with different medication profiles) was measured in a genome-wide approach to characterize the late fluoxetine-induced gene expression changes and to identify potential gene expression biomarkers. Gene expression profiles were compared between untreated samples and samples treated for 21 days with 0.5 μg ml−1 of fluoxetine, which is similar to the average plasma concentration in fluoxetine-medicated patients. The responder- and non-responder-derived indicator cell lines were compared (characterized by rectified clinical response status, experimental EdU proliferation status and hierarchical clustering status obtained from microarray experiments), seven inversely regulated pathways were highlighted containing a total of 192 differentially expressed genes after incubation with fluoxetine (fold change >2, P-value <0.05). In those cell lines, STRING-based gene ontology analysis revealed 127 of the identified genes as being involved in brain remodeling (Table 2 and Supplementary Table 3). After consideration of LCL donor’s individual response status (cell lines derived from n=6 responders vs n=4 non-responders), 15 genes showed different expression between responder- and non-responder-derived cell lines and, therefore, were used for further investigation and were considered as candidate genes. The gene names, fold-change differences, as well as their annotated central nervous system functions are listed in Table 3.
Cell proliferation
To assess the individual differences in cell proliferative effects of fluoxetine in LCLs from patients with documented clinical response status, we conducted long-term cell incubation with fluoxetine revealing large variability in relative proliferation rates ranging from 55 to 155% in comparison with untreated cells from the same donor (Figure 2a). The covariates age and gender showed no significant impact on individual proliferation rates (Figure 2b). When grouping the cell lines according to their donor’s clinical response status, no significant differences between the proliferation rates of the single groups were detectable (Figure 2c). Furthermore, no association was detected between LCL proliferation rates and LCL donor’s clinical response measured as percentage change in Hamilton score compared between weeks 0 and 8 (Figure 2d).
Results from EdU (5-ethynyl-2'-deoxyuridine) phenotyping experiments show individual differences between the cell lines (a). The covariates gender and age do not significantly influence the individual proliferation rates (b). Box plot analysis of EdU cell proliferation reveals no significant difference in proliferation rates after consideration of clinical response/remission status after 5 and 8 weeks of treatment (c). Detailed overview on the correlation between relative proliferation rates and response factor (defined as percentage change in Hamilton score compared between week 0 and 8). Each dot represents results from one cell line. As indicated by the trend line, no association between proliferation rates and response status is recognizable (d).
Real-time gene expression analyses of the candidate genes
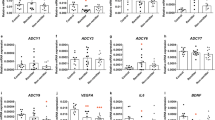
To assess the potential of the identified candidate genes, gene expression was analyzed in an edge-group approach similar to the work of Morag et al.10 From the two phenotypic edges of EdU phenotyping (five cell lines each)—those cell lines with the most distinct fluoxetine-induced anti-proliferative and pro-proliferative effects—basal gene expression and fluoxetine-induced changes were compared. Among the 15 identified genes from our microarray experiments, the basal gene expression of four genes was significantly different from proliferator cell lines compared with non-proliferator cell lines: wingless-type MMTV integration site family, member 2B (WNT2B), transcription factor 7-like 2 (TCF7L2) sulfotransferase 4A1 (SULT4A1) and P-glycoprotein (ABCB1; Figure 3). After consideration of the LCL donor’s clinical response status, no significant differences between gene expression of non-responder- and responder-derived LCLs were detectable (data not shown). In several cell lines, fluoxetine-induced gene expression changes of the above genes as well as FZD7 (frizzled class receptor 7) were observed. Results of the fold-change analyses significantly correlated with in vitro proliferation of genes WNT2B, TCF7L2 and FZD7 (Table 4).
Changes in candidate gene expression were assessed after 21 days incubation with fluoxetine in all LCLs from EdU phenotyping experiments (n=50). The associations between gene expression, and both the remission and response status of LCL donors were investigated. Basal gene expression of SULT4A1 correlated with clinical response after 5 weeks (P=0.029). However, basal gene expression of SULT4A1 was low, and only detectable in 10 (n=4 non-responder-derived cell lines vs n=6 responder-derived cells) out of 50 cell lines. Furthermore, the gene expression fold-change values of WNT2B after treatment with fluoxetine correlated with LCL donor’s clinical remission status after 5 weeks (P=0.025). The remaining genes TCF7L2, FZD7 and ABCB1 showed no significant correlations with clinical parameters of LCL donors (see Supplementary Table 4).
Discussion
Peripheral proliferation is unsuitable as surrogate marker for antidepressant response
In search of tentative functional biomarkers for antidepressant response prediction, we tested fluoxetine effects on cell proliferation in LCLs from depressed patients. Individual effects on cell proliferation have been detected after 21 days of incubation with fluoxetine. Although the in vitro treatment of patient-derived LCLs with fluoxetine presents high inter-individual variability regarding the LCL proliferation behavior, this phenomenon has—according to our data—no association with the patient’s clinical outcome.
Our initial hypothesis was based on the assumption that antidepressants induce the proliferation of neuronal cells and therefore modulate the neural plasticity.3 Depressed patients show a volume reduction of depression-associated brain parts,39 that might be reversed by antidepressant-induced proliferation.6 The stimulation of neuronal stem cell proliferation in the brain is directly linked with an enhanced neuroplasticity, which eventually leads to a normalization of the depressed mood.7 As cerebral remodeling processes are complex and take many weeks, this phenomenon explains the observed delay (from weeks up to several months) in symptomatic improvement. Nonetheless, direct proliferative effects of antidepressants were observed (for example, by Chang et al.19) by several research groups in rodents40, 41 and non-human primates42 and by Chen et al.43 in a genetic rat model of depression. The molecular mechanisms underlying remission of depression remain unclear, although neurotrophic growth factors—like brain-derived neurotrophic factor—may have an important role during remission processes.44 One reason for the lack in association between clinical response and in vitro cell proliferation effects of in vitro fluoxetine treatment may be that the cell model in blood-derived LCLs is not suitable for studying brain-specific antidepressant-induced proliferative effects owing to lack of relevant neuronal pathways. Yet, several observations support a potential role of LCLs for the study of tentative biomarkers for individual variability of drug effects. For example, Morag et al.10 identified different neuronal genes (for example, CHL1, ITGB3 or GAP43) as potential gene expression biomarkers to predict the response based on individual paroxetine sensitivity in LCLs. A further study using LCLs derived from depressed patients confirmed some of these genes as potential gene expression biomarkers for the prediction of individual antidepressant response.45 In another study, they presented a LCL-based tool to assess shared drug pathways, that was developed by comparing growth-inhibition profiles of different drug classes (including antidepressants) and can be used to categorize distinct pathways.12 Oved et al.46 identified potential antidepressant drug targets by genome-wide expression profiling and tentative response biomarkers in human LCLs.9 In addition to these studies that focused on LCLs as tools for the identification of biomarkers for depressive disorders, a few studies explored the utility of LCLs in other psychological diseases such as bipolar disorders or autism.47, 48, 49
Moreover, single-nucleotide polymorphisms in neuronal cell adhesion genes involved in synaptic plasticity and identified in the two latter studies performed with human LCLs, namely, CHL1 and ITGB3, were recently shown to affect treatment response in depressive disorders.50
Identification of potential gene expression biomarkers
Phenotyping the proliferative response of LCLs to fluoxetine (0.5 μg ml−1; 21 days) followed by comparative microarray-based genome-wide gene expression profiling revealed candidate genes being involved in brain remodeling processes. Genome-wide analyses of fluoxetine-induced gene expression changes in human LCLs from patients with characterized antidepressant drug response resulted in significant transcriptional regulation of 15 genes involved in neurogenesis. As microarray analyses were slightly underpowered and uncorrected for multiple testing (no false discovery rate correction), the results from the microarray analyses were further validated by RT-PCR (in the edge-group approach and—the remaining candidate genes—in all n=50 cell lines). By far, the strongest gene expression differences compared between responder-derived cell lines relative to non-responder-derived cell lines were obtained for betacellulin (BTC; with the following mean fold changes: +40.0 in responder-derived cell lines and −0.3 in non-responder-derived cell lines). BTC belongs to the EGF (epidermal growth factor) protein family and has been reported to stimulate neurogenesis,26 as well as neural stem cell proliferation and differentiation into glial- and neuronal-like cell types.51 BTC is endogenously produced in the brain, especially by blood vessels and the choroid plexus, and directly affects neuroblast differentiation and neuronal stem cell regeneration by activation of EGFR and ERBB4. It is considered a potential therapeutic agent for treating neurodegenerative diseases.26
Five genes (WNT2B, TCF7L2, FZD7, SULT4A1 and ABCB1) were differently expressed in cell lines with the highest increase vs highest decrease in cell proliferation following 21 days fluoxetine incubation. Data analysis showed a correlation between LCL donor’s clinical response (in n=6 responders and n=4 non-responders) and the basal gene expression of SULT4A1. Furthermore, the gene expression fold changes of WNT2B by fluoxetine incubation correlated with LCL donor’s clinical remission. None of the remaining genes TCF7L2, FZD7 and ABCB1 showed significant correlation with clinical parameters of LCL donors.
The transcription factor TCF7L2 and the receptor FZD7 belong to the WNT signaling pathway, which has an important role for regulation of stem cell pluripotency and cell differentiation by integrating signals from other pathways and their associated signal molecules such as fibroblast growth factor52 and bone morphogenic protein.53 Both growth factors are involved in depression pathogenesis54, 55 and in the maintenance of adult hippocampal neurogenesis (together with brain-derived neurotrophic factor, vascular endothelial growth factor and other signaling pathways).56 WNT2B belongs to a family of highly conserved signal molecules involved in the regulation of neural cell growth and differentiation.27 Furthermore, Wnt signaling regulates adult hippocampal neurogenesis57 and the expansion of central nervous system progenitor cells.58 Moreover, it is important for synaptic function as well as for the formation of hippocampal spines.59, 60 A malfunction of Wnt signaling in the hippocampus by targeted knockdown is associated with decreased neurogenesis, increased depression-like behavior and various neuropsychiatric disorders.61, 62 Wnt glycoproteins are released by hippocampal astrocytes and take effects through gene expression activation of NeuroD and Dcx,63, 64 a transcription factor involved in central nervous system development and a microtubule-associated protein almost exclusively expressed in actively dividing neuronal precursor cells, respectively.65, 66 It has been shown that Wnt signaling is responsive to various antidepressant drugs,67 whereas mice with constitutively activated Wnt signaling become irresponsive to antidepressant treatments.68 Furthermore, a role of Wnt signaling via the fast-acting antidepressant ketamine has been proposed.69
Little is known so far about the brain-specific phase II metabolizing enzyme SULT4A1, but it may be involved in the metabolism of antidepressant drugs and neuroactive substances.30 However, as expression of SULT4A1 was low in LCLs and only detectable in 10 out of 50 cell lines, these results should be seen with caution and warrants further analysis, of SULT4A1 expression in brain. The transporter ABCB1 belongs to the ATP-binding cassette superfamily possessing a key role in transmembrane transport. ABCB1 is an efflux pump with a broad substrate spectrum (including a variety of antidepressant drugs as well as neurotoxic agents) transporting these substances through the blood–brain barrier into the circulatory system. The resulting ABCB1-mediated neuroprotective effect might contribute to an increased proliferation of neuronal cells. Several single-nucleotide polymorphisms in the ABCB1 gene were associated with depression severity, response status or dosage adjustments in depressive disorders indicating an involvement of ABCB1 in depression.70, 71, 72, 73 Fluoxetine, the antidepressant we used for in vitro LCL phenotyping, is a rather weak substrate of ABCB1.74, 75 One may speculate that the absence of a correlation between fluoxetine-induced ABCB1 expression and clinical response could reflect the low ABCB1 substrate properties of this antidepressant drug.
Conclusion
We measured proliferative effects of fluoxetine (21 days) in LCLs from depressed patients and analyzed association between gene expression changes of the genes identified by microarray analysis and cell proliferation. Three of the 15 genes identified from genome-wide analyses showed significant associations with cell proliferative behavior. Furthermore, for the gene expression of two candidate genes, SULT4A1 and WNT2B, we observed correlations between LCL donor’s clinical response and remission, respectively. These genes are involved in the metabolism of antidepressants and neuroactive agents, and in neural cell proliferation and differentiation, respectively. Further studies should follow to elucidate a connection between cellular proliferation effects and clinical antidepressant response. The candidate genes reported here should be further examined for their pharmacogenetic variability and their role in remission from depression using longitudinal blood samples from major depression patients, as well as brain tissues from animal models for depression.
Accession codes
References
Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, STEWART JW et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: A STAR*D report. Am J Psychiatry 2006; 163: 13.
Hennings JM, Owashi T, Binder EB, Horstmann S, Menke A, Kloiber S et al. Clinical characteristics and treatment outcome in a representative sample of depressed inpatients – Findings from the Munich Antidepressant Response Signature (MARS) project. J Psychiatr Research 2009; 43: 215–229.
Castrén E, Hen R . Neuronal plasticity and antidepressant actions. Trends Neurosci 2013; 36: 259–267.
Mahar I, Bambico FR, Mechawar N, Nobrega JN . Stress, serotonin, and hippocampal neurogenesis in relation to depression and antidepressant effects. Neurosci Biobehav Rev 2014; 38: 173–192.
Frodl T, Meisenzahl EM, Zetzsche T, Born C, Groll C, Jäger M et al. Hippocampal changes in patients with a first episode of major depression. Am J Psychiatry 2002; 159: 1112–1118.
Malberg JE, Duman RS . Cell proliferation in adult hippocampus is decreased by inescapable stress: reversal by fluoxetine treatment. Neuropsychopharmacology 2003; 28: 1562–1571.
Pilar-Cuellar F, Vidal R, Diaz A, Castro E, dos Anjos S, Pascual-Brazo J et al. Neural plasticity and proliferation in the generation of antidepressant effects: hippocampal implication. Neural Plast 2013; 2013: 21.
Tanti A, Belzung C . Hippocampal neurogenesis: a biomarker for depression or antidepressant effects? Methodological considerations and perspectives for future research. Cell Tissue Res 2013; 354: 203–219.
Oved K, Morag A, Pasmanik-Chor M, Oron-Karni V, Shomron N, Rehavi M et al. Genome-wide miRNA expression profiling of human lymphoblastoid cell lines identifies tentative SSRI antidepressant response biomarkers. Pharmacogenomics 2012; 13: 1129–1139.
Morag A, Pasmanik-Chor M, Oron-Karni V, Rehavi M, Stingl JC, Gurwitz D . Genome-wide expression profiling of human lymphoblastoid cell lines identifies CHL1 as a putative SSRI antidepressant response biomarker. Pharmacogenomics 2011; 12: 171–184.
Cobb JA, Simpson J, Mahajan GJ, Overholser JC, Jurjus GJ, Dieter L et al. Hippocampal volume and total cell numbers in major depressive disorder. J Psychiatr Res 2013; 47: 299–306.
Morag A, Kirchheiner J, Rehavi M, Gurwitz D . Human lymphoblastoid cell line panels: novel tools for assessing shared drug pathways. Pharmacogenomics 2010; 11: 327–340.
Ising M, Lucae S, EBa Binder, Bettecken T, Uhr M, Ripke S et al. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry 2009; 66: 966–975.
Ising M, Horstmann S, Kloiber S, Lucae S, Binder EB, Kern N et al. Combined dexamethasone/corticotropin releasing hormone test predicts treatment response in major depression–a potential biomarker? Biol Psychiatry 2007; 62: 47–54.
Hamilton M . A rating scale for depression. J Neurol Neurosurg Psychiatry 1960; 23: 56–62.
Zobel AW, Nickel T, Sonntag A, Uhr M, Holsboer F, Ising M . Cortisol response in the combined dexamethasone/CRH test as predictor of relapse in patients with remitted depression: a prospective study. J Psychiatr Res 2001; 35: 83–94.
Anderson MA, Gusella JF . Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In Vitro 1984; 20: 856–858.
Tosato G, Cohen JI . Generation of Epstein-Barr Virus (EBV)–immortalized B cell lines. Curr Protoc Immunol 2007; Chapter 7: Unit 7.22.
Chang E-A, Beyhan Z, Yoo M-S, Siripattarapravat K, Ko T, Lookingland KJ et al. Increased cellular turnover in response to fluoxetine in neuronal precursors derived from human embryonic stem cells. Int J Dev Biol 2010; 54: 707–715.
Sousa-Ferreira L, Aveleira C, Botelho M, Álvaro A, Pereira de Almeida L, Cavadas C . Fluoxetine induces proliferation and inhibits differentiation of hypothalamic neuroprogenitor cells in vitro. PLoS One 2014; 9: e88917.
Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science 2003; 301: 805–809.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43 (Database issue): D447–D452.
Edgar R, Domrachev M, Lash AE . Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30: 207–210.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 2013; 41: D991–D995.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001; 25: 402–408.
Gómez-Gaviro MV, Scott CE, Sesay AK, Matheu A, Booth S, Galichet C et al. Betacellulin promotes cell proliferation in the neural stem cell niche and stimulates neurogenesis. Proc Natl Acad Sci USA 2012; 109: 1317–1322.
Kubo F, Takeichi M, Nakagawa S . Wnt2b inhibits differentiation of retinal progenitor cells in the absence of Notch activity by downregulating the expression of proneural genes. Development 2005; 132: 2759–2770.
Aguirre A, Rubio ME, Gallo V . Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature 2010; 467: 323–327.
Brandl E, Chowdhury N, Tiwari A, Lett TP, Meltzer H, Kennedy J et al. Genetic variation in CYP3A43 is associated with response to antipsychotic medication. J Neural Transm (Vienna) 2015; 122: 29–34.
Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A et al. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol 2007; 5: e97.
Wu Melody V, Hen R . The young and the restless: regulation of adult neurogenesis by Wnt signaling. Cell Stem Cell 2013; 12: 139–140.
Boehm C, Newrzella D, Herberger S, Schramm N, Eisenhardt G, Schenk V et al. Effects of antidepressant treatment on gene expression profile in mouse brain: cell type-specific transcription profiling using laser microdissection and microarray analysis. J Neurochem 2006; 97: 44–49.
Hayashi H . Lipid metabolism and glial lipoproteins in the central nervous system. Biol Pharm Bull 2011; 34: 453–461.
Tarr PT, Edwards PA . ABCG1 and ABCG4 are coexpressed in neurons and astrocytes of the CNS and regulate cholesterol homeostasis through SREBP-2. J Lipid Res 2008; 49: 169–182.
Puschmann TB, Zandén C, Lebkuechner I, Philippot C, de Pablo Y, Liu J et al. HB-EGF affects astrocyte morphology, proliferation, differentiation, and the expression of intermediate filament proteins. J Neurochem 2014; 128: 878–889.
Ries V, Silva RM, Oo TF, Cheng H-C, Rzhetskaya M, Kholodilov N et al. JNK2 and JNK3 combined are essential for apoptosis in dopamine neurons of the substantia nigra, but are not required for axon degeneration. J Neurochem 2008; 107: 1578–1588.
Schinkel AH . P-Glycoprotein, a gatekeeper in the blood–brain barrier. Adv Drug Deliv Rev 1999; 36: 179–194.
Britsch S, Li L, Kirchhoff S, Theuring F, Brinkmann V, Birchmeier C et al. The ErbB2 and ErbB3 receptors and their ligand, neuregulin-1, are essential for development of the sympathetic nervous system. Genes Dev 1998; 12: 1825–1836.
aan het Rot M, Mathew SJ, Charney DS . Neurobiological mechanisms in major depressive disorder. CMAJ 2009; 180: 305–313.
Wang J-W, David DJ, Monckton JE, Battaglia F, Hen R . Chronic fluoxetine stimulates maturation and synaptic plasticity of adult-born hippocampal granule cells. J Neurosci 2008; 28: 1374–1384.
Khemissi W, Farooq RK, Le Guisquet A-M, Sakly M, Belzung C . Dysregulation of the hypothalamus-pituitary-adrenal axis predicts some aspects of the behavioral response to chronic fluoxetine: association with hippocampal cell proliferation. Front Behav Neurosci 2014; 8: 340.
Perera TD, Coplan JD, Lisanby SH, Lipira CM, Arif M, Carpio C et al. Antidepressant-induced neurogenesis in the hippocampus of adult nonhuman primates. J Neurosci 2007; 27: 4894–4901.
Chen F, Madsen TM, Wegener G, Nyengaard JR . Imipramine treatment increases the number of hippocampal synapses and neurons in a genetic animal model of depression. Hippocampus 2010; 20: 1376–1384.
Yu H, Chen Z-Y . The role of BDNF in depression on the basis of its location in the neural circuitry. Acta Pharmacol Sin 2011; 32: 3–11.
Probst-Schendzielorz K, Scholl C, Efimkina O, Ersfeld E, Viviani R, Serretti A et al. CHL1, ITGB3 and SLC6A4 gene expression and antidepressant drug response: results from the Munich Antidepressant Response Signature (MARS) study. Pharmacogenomics 2015; 16: 689–701.
Oved K, Morag A, Pasmanik-Chor M, Rehavi M, Shomron N, Gurwitz D . Genome-wide expression profiling of human lymphoblastoid cell lines implicates integrin beta-3 in the mode of action of antidepressants. Transl Psychiatry 2013; 3: e313.
Squassina A, Costa M, Congiu D, Manchia M, Angius A, Deiana V et al. Insulin-like growth factor 1 (IGF-1) expression is up-regulated in lymphoblastoid cell lines of lithium responsive bipolar disorder patients. Pharmacol Res 2013; 73: 1–7.
Chen H, Wang N, Burmeister M, McInnis MG . MicroRNA expression changes in lymphoblastoid cell lines in response to lithium treatment. Int J Neuropsychopharmacol 2009; 12: 975.
Hu VW, Frank BC, Heine S, Lee NH, Quackenbush J . Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics 2006; 7: 118.
Fabbri C, Crisafulli C, Gurwitz D, Stingl J, Calati R, Albani D et al. Neuronal cell adhesion genes and antidepressant response in three independent samples. Pharmacogenomics J 2015; 15: 538–548.
McKiernan E, O'Driscoll L, Kasper M, Barron N, O'Sullivan F, Clynes M . Directed differentiation of mouse embryonic stem cells into pancreatic-like or neuronal-and glial-like phenotypes. Tissue Eng 2007; 13: 2419–2430.
Israsena N, Hu M, Fu W, Kan L, Kessler JA . The presence of FGF2 signaling determines whether β-catenin exerts effects on proliferation or neuronal differentiation of neural stem cells. Dev Biol 2004; 268: 220–231.
Kléber M, Lee H-Y, Wurdak H, Buchstaller J, Riccomagno MM, Ittner LM et al. Neural crest stem cell maintenance by combinatorial Wnt and BMP signaling. J Cell Biol 2005; 169: 309–320.
Evans SJ, Choudary PV, Neal CR, Li JZ, Vawter MP, Tomita H et al. Dysregulation of the fibroblast growth factor system in major depression. Proc Natl Acad Sci USA 2004; 101: 15506–15511.
Ordway GA, Szebeni A, Chandley MJ, Stockmeier CA, Xiang L, Newton SS et al. Low gene expression of bone morphogenetic protein 7 in brainstem astrocytes in major depression. Int J Neuropsychopharmacol 2012; 15: 855–868.
Fuentealba Luis C, Obernier K, Alvarez-Buylla A . Adult neural stem cells bridge their niche. Cell Stem Cell 2012; 10: 698–708.
Lie D-C, Colamarino SA, Song H-J, Desire L, Mira H, Consiglio A et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature 2005; 437: 1370–1375.
Ikeya M, Lee SMK, Johnson JE, McMahon AP, Takada S . Wnt signalling required for expansion of neural crest and CNS progenitors. Nature 1997; 389: 966–970.
Sahores M, Gibb A, Salinas PC . Frizzled-5, a receptor for the synaptic organizer Wnt7a, regulates activity-mediated synaptogenesis. Development 2010; 137: 2215–2225.
Varela-Nallar L, Grabowski CP, Alfaro IE, Alvarez AR, Inestrosa NC . Role of the Wnt receptor Frizzled-1 in presynaptic differentiation and function. Neural Dev 2009; 4: 41.
Inestrosa NC, Arenas E . Emerging roles of Wnts in the adult nervous system. Nat Rev Neurosci 2010; 11: 77–86.
Jessberger S, Clark RE, Broadbent NJ, Clemenson GD, Consiglio A, Lie DC et al. Dentate gyrus-specific knockdown of adult neurogenesis impairs spatial and object recognition memory in adult rats. Learn Mem 2009; 16: 147–154.
Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, Lie DC et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci 2009; 12: 1097–1105.
Vanderhaeghen P . Wnts blow on NeuroD1 to promote adult neuron production and diversity. Nat Neurosci 2009; 12: 1079–1081.
Bayatti N, Sarma S, Shaw C, Eyre JA, Vouyiouklis DA, Lindsay S et al. Progressive loss of PAX6, TBR2, NEUROD and TBR1 mRNA gradients correlates with translocation of EMX2 to the cortical plate during human cortical development. Eur J Neurosci 2008; 28: 1449–1456.
Couillard-Despres S, Winner B, Schaubeck S, Aigner R, Vroemen M, Weidner N et al. Doublecortin expression levels in adult brain reflect neurogenesis. Eur J Neurosci 2005; 21: 1–14.
Okamoto H, Voleti B, Banasr M, Sarhan M, Duric V, Girgenti MJ et al. Wnt2 expression and signaling is increased by different classes of antidepressant treatments. Biol Psychiatry 2010; 68: 521–527.
Eom T-Y, Jope RS . Blocked inhibitory serine-phosphorylation of glycogen synthase kinase-3α/β impairs in vivo neural precursor cell proliferation. Biol Psychiatry 2009; 66: 494–502.
Machado-Vieira R, Salvadore G, DiazGranados N, Zarate CA . Ketamine and the next generation of antidepressants with a rapid onset of action. Pharmacol Ther 2009; 123: 143–150.
Lin K-M, Chiu Y-F, Tsai I-J, Chen C-H, Shen WW, Liu SC et al. ABCB1 gene polymorphisms are associated with the severity of major depressive disorder and its response to escitalopram treatment. Pharmacogenet Genomics 2011; 21: 163–170.
Xie W-W, Zhang L, Wu R-R, Yu Y, Zhao J-P, Li L-H . Case-control association study of ABCB1 gene and major depressive disorder in a local Chinese Han population. Neuropsychiatr Dis Treat 2015; 11: 1967–1971.
Singh AB, Bousman CA, Ng CH, Byron K, Berk M . ABCB1 polymorphism predicts escitalopram dose needed for remission in major depression. Transl Psychiatry 2012; 2: e198.
Uhr M, Tontsch A, Namendorf C, Ripke S, Lucae S, Ising M et al. Polymorphisms in the drug transporter gene ABCB1 predict antidepressant treatment response in depression. Neuron 2008; 57: 203–209.
Uhr M, Steckler T, Yassouridis A, Holsboer F . Penetration of amitriptyline, but not of fluoxetine, into brain is enhanced in mice with blood-brain barrier deficiency due to mdr1a P-glycoprotein gene disruption. Neuropsychopharmacology 2000; 22: 380–387.
Doran A, Obach RS, Smith BJ, Hosea NA, Becker S, Callegari E et al. The impact of P-glycoprotein on the disposition of drugs targeted for indications of the central nervous system: evaluation using the MDR1A/1B knockout mouse model. Drug Metab Dispos 2005; 33: 165–174.
Acknowledgements
The cooperation with the Munich MARS study, and generation of LCLs from blood of the patients was supported by financial support from the ERANET project PADRE on antidepressant drug response biomarkers (JCS, 01EW1105).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
FH is the founder and CEO of HMNC Brain Health GmbH Munich and holds two patents for the prediction of antidepressant treatment response by the ABCB1 gene (WO 2005/108605: Polymorphisms in ABCB1 associated with a lack of clinical response to medicaments; WO 2008/151803: New polymorphisms in ABCB1 associated with a lack of clinical response to medicaments). No writing assistance was utilized in the production of this manuscript. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Breitfeld, J., Scholl, C., Steffens, M. et al. Proliferation rates and gene expression profiles in human lymphoblastoid cell lines from patients with depression characterized in response to antidepressant drug therapy. Transl Psychiatry 6, e950 (2016). https://doi.org/10.1038/tp.2016.185
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2016.185
This article is cited by
-
Potential depression and antidepressant-response biomarkers in human lymphoblast cell lines from treatment-responsive and treatment-resistant subjects: roles of SSRIs and omega-3 polyunsaturated fatty acids
Molecular Psychiatry (2021)
-
Citalopram-induced pathways regulation and tentative treatment-outcome-predicting biomarkers in lymphoblastoid cell lines from depression patients
Translational Psychiatry (2020)
-
The metabolic network coherence of human transcriptomes is associated with genetic variation at the cadherin 18 locus
Human Genetics (2019)
-
Gene expression and proliferation biomarkers for antidepressant treatment resistance
Translational Psychiatry (2017)