Abstract
Non-alcoholic fatty liver disease (NAFLD) is characterized by excessive lipid accumulation in hepatocytes. The role of SENP3 in lipid metabolism, particularly NAFLD, is unclear. Our results showed that hepatic SENP3 was up-regulated in NAFLD patients and an animal model in vivo and after loading hepatocytes with free fatty acids (FFA) in vitro. Intracellular lipid accumulation was determined in SENP3 silenced or overexpressed hepatocytes with/without FFA in vitro. Confirming a role for SENP3, gene silencing was associated in vitro with amelioration of lipid accumulation and overexpression with enhancement of lipid accumulation. SENP3 related genes in NAFLD were determined in vitro using RNA-Seq. Eleven unique genes closely associated with lipid metabolism were generated using bioinformatics. Three selected genes (apoe, a2m and tnfrsf11b) were verified in vitro, showing apoe, a2m and tnfrsf11b were regulated by SENP3 with FFA stimulation. Intrahepatic and circulating APOE, A2M and TNFRSF11B were elevated in NAFLD compared with controls. These data demonstrate the important role of SENP3 in lipid metabolism during the development of NAFLD via downstream genes, which may be useful information in the development of NAFLD. The precise role of SENP3 in NAFLD will be investigated using liver-specific conditional knockout mice in future studies.
Similar content being viewed by others
Introduction
Non-alcoholic fatty liver disease (NAFLD), due to aberrant lipid metabolism, has become an increasingly prevalent health problem worldwide1. Anomalous lipid metabolism plays a crucial role in the progression of NAFLD, attacking hepatocytes2. Excessive accumulation of triglyceride (TG) in hepatocytes, as a precursor for NAFLD, is due to increased uptake of free fatty acids (FFA) into the liver, up-regulated triglyceride synthesis, impaired lipid β-oxidation, or weakened VLDL secretion3. Excessive TG is stored in hepatocytes as lipid droplets. Perturbed metabolic pathways (e.g. mTOR and AKT/PI3K signaling pathway) and abnormal expression of related molecules [e.g. Apo lipoproteins, free acid binding proteins (FABPs), liver X receptors] contribute to the formation of lipid droplets in the onset of steatosis in NAFLD4. Hepatic steatosis increases the risk of irreversible liver disease, ultimately leading to liver fibrosis, cirrhosis or hepatocellular carcinoma5.
Ubiquitin-like SUMO (small ubiquitin-related modifiers), including SUMO1, SUMO2 and SUMO3, are involved in post-translational modification of target proteins. SUMOs regulate a broad spectrum of cellular processes, e.g. cell signaling, cycle, transcription, or carcinogenesis6. SUMOylation is a dynamic process that can be reversed by a family of SUMO-specific proteases (SENPs)7. Six SENP members in mammals, identified with different subcellular locations and substrates, play important roles in the control of various cellular events7. SENPs, by acting in the protein regulatory event deSUMOylation, play important roles during the progression of many diseases. It has been reported that overexpressed SENP1 promotes mitochondrial biogenesis and function in myotubes8. SENP2 inhibits glycolysis, reprograms glucose metabolic strategy in cancer cells9 and regulates fatty acid metabolism in skeletal muscle10, suggesting an important role of SENP2 in metabolic disorders. SENP5 is reported to maintain mitochondrial morphology and metabolism11. As a crucial member of the SENPs, SENP3 is specific to de-conjugate SUMO2 or SUMO3, which share 96% sequence similarity12. SENP3 is rapidly increased following several cell stresses e.g. oxidative stress or hyperglycemic stress13,14. SENP3 is up-regulated in the progression of many diseases, including cancers13 and other injuries15. Up-regulated SENP3 enhances cell apoptosis/death16, proliferation13 and cancer metastasis17. However, it is unknown whether SENP3 and SENP3-related molecules contribute to the progression of NAFLD via manipulation of lipid accumulation in hepatocytes. Thus, the aim of the current study is to explore the role of SENP3 in the development of NAFLD.
Materials and Methods
Ethical considerations
All the experiments protocols involving humans and animals were approved by the Human Ethics Committee of Ruijin Hospital, Shanghai Jiaotong University School of Medicine. Methods were carried out in accordance with the approved guidelines and regulation. Written informed consent was obtained from all participants. Appropriate care was given to all animals included for experiments.
Patients
Plasma from NAFLD patients (n = 50) and healthy controls (HCs) (n = 40) [2012 AASLD criteria5], identified between Jan 2014 and Oct 2015 in the Department of Infectious Diseases, Ruijin Hospital, Shanghai, China was collected and stored at −80 °C before analysis. Liver biopsies from NAFLD patients (n = 8) and liver transplant donors (n = 3) were also collected in the Department of Infectious Diseases, Ruijin Hospital, Shanghai, China. Paraffin embedded liver tissues were used for immunohistochemistry (IHC).
Animal studies
Five-week-old male SD rats, each weighing 150–160 g were purchased from the Shanghai Laboratory Animal Company (Shanghai, China). All rats were bred in a specific pathogen-free facility and maintained in a 12-hour light-dark-cycle at room temperature and fed ad libitum. The rats were randomly divided into two groups, normal diet (ND) fed with standard chow (n = 5) or high fat diet (HFD) fed with 45% fat chow (n = 5)18. The rats were sacrificed on the 60th day. Serum was collected and stored at −80 °C before analysis. Part of the liver tissues were fixed with 10% formalin and embedded in paraffin for routine HE staining and IHC, while other parts were snap frozen and stored in liquid nitrogen until use. Hepatic TG content was determined using the TG assay kit (Applygen Technologies Inc., Beijing, China). In addition, livers from HFD fed 30, 60 and 120 day rats (n = 5, respectively) were collected for Western blotting, and the correlation between hepatic SENP3 and TG was determined.
Cell culture
Human normal hepatocytes (L02 cell line), obtained from China Cell Culture Center (Shanghai, China), were cultured in Gibco® RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (Sigma, MO, USA) under an atmosphere of 5% CO2 at 37 °C. Hepatocytes were exposed to free fatty acid (FFA), a mixture of oleic acid and palmitic acid (Sigma, MO, USA), at the final ratio of 2:1, with a total concentration of 0.5 mmol/L for 12 hours to establish steatotic hepatocytes. SENP3-siRNA (siRNA specific for SENP3) and NS-siRNA (non-specific siRNA) oligonucleotides were purchased from Ribobio (Guangzhou, China). The sequences of SENP3-siRNA oligonucleotides were 5′- GGGCUGGAAAGGUUACUUCdTdT-3′ and 3′-dTdTCCCGACCUUUCCAAUGAAG-5′. GFP-SENP3 plasmid and PCDNA3.1 (vehicle plasmid) were kind gifts from the Education Ministry Key Laboratory for Cell Differentiation and Apoptosis, Shanghai Jiaotong University School of Medicine, China13,19. These hepatocytes were transfected with the siRNA oligonucleotides or plasmids using lipofectamine 2000 (Invitrogen, USA), according to the manufacturer’s instructions. The transfected hepatocytes were harvested with/without 12 hours FFA stimulation for quantification of intracellular lipid accumulation using Oil red O (Sigma, USA) staining20, or intracellular TG content using the TG assay kit (Applygen Technologies Inc.).
RNA-Seq (mRNA sequencing)
Total RNA was extracted from the three hepatocyte treatment groups: FAA treated only, SENP3 silenced with FFA treatment, and SENP3 overexpressed with FFA treatment. Total RNA was isolated using the standard Trizol method and paired-end library preparations were performed. RNA-Seq was performed to determine the mRNA expression profile using a large scale, automated variant of the Illumina HiSeq 2500 v4 (Berry Genomics Co., Ltd, Beijing, China). All the clean reads after raw data pre-processing were aligned against the reference genome and transcriptome data downloaded from UCSC (version hg19), using the SOAP2 package21. The reads uniquely mapped to a gene to quantified transcript levels were normalized by the RPKM method22.
Analysis of differentially expressed genes (DEGs)
DEGs were identified using the fold change method and adjusted by the generalized fold change algorithm (GFOLD)23 between two groups. To identify DEGs, the cut-off value with absolute GFOLD value of 1 was selected. All the scripts used perl and R. All the DEGs were performed against the ExoCarta database24 to obtain secreted proteins in plasma or serum. To enrich the lipid related metabolism pathways, all secreted DEGs were evaluated, using the Ingenuity Pathway Analysis (IPA) (Ingenuity® Systems, www.ingenuity.com) against human species. The gene list was generated from the IPA result associated with lipid metabolism and narrowed down with disease functions of steatosis and steatohepatitis.
Western blotting
The protein concentration was measured following total protein extraction from hepatic tissues or cultured hepatocytes, using RIPA lysis buffer (Beyotime, China). Western blotting was performed, using antibodies against SENP3 (Cell Signaling Technology, MA, USA) and β-actin (Sigma, USA). Protein production was quantitated using Image J.
Quantitative real-time PCR (qRT-PCR)
qRT-PCR on select genes were performed to verify the bioinformatics derived from RNAseq, as described25 using SYBR® Premix Ex TaqTM II (Takara, Dalian, China) and an ABI ViiA7 instrument (Applied Biosystems, USA). Relative expression levels of mRNA were calculated by applying the 2−ΔΔCt method using reference gapdh for mRNA and normalized to the control group. Sequences of primers used are outlined in Supplementary Materials and Methods.
Immunohistochemistry (IHC)
Five μm sections of the liver tissues were labelled immunohistochemically and quantitated using Image-Pro Plus 9.0 software (Diagnostic Instruments, Sydney, NSW, Australia) and the data were expressed as image units, as described previously26. The primary antibody against SENP3 was purchased from Cell Signaling Technology, and the primary antibodies against APOE, A2M and TNFRSF11B (also named Osteoprotegerin, OPG) were purchased from Abcam, Cambridge, MA, USA.
ELISA
Circulating levels of A2M, APOE and TNFRSF11B were measured according to the instructions from the manufacturers (Abcam, MA, USA).
Statistical analysis
Values were presented as mean ± SEM. All statistical analyses were carried out using GraphPad Prism 5 and Sigmaplot 12.5 for Windows. Statistically significance was regarded as *p < 0.05, **p < 0.01 and ***p < 0.001.
Results
SENP3 was significantly up-regulated in the livers from NAFLD patients and HFD fed rats
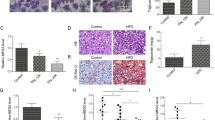
To investigate if SENP3 was modified in NAFLD, the production of SENP3 was examined in livers from liver transplant donors and NAFLD patients. Histopathology of the liver tissues from liver donors (Fig. 1A) and NAFLD (Fig. 1B), and immunohistochemistry for SENP3 in livers from liver donors (Fig. 1C) and NAFLD (Fig. 1D) were stained. SENP3 was mainly localized around the nuclei of hepatocytes, consistent with high levels of fatty deposition. The relative mean density of SENP3 was ~20 fold higher in the livers from NAFLD than that in livers from donors (Fig. 1E, p < 0.001), suggesting the importance of SENP3 during the development of NAFLD. Due to lack of sufficient human liver tissue for further confirmation of SENP3 for Western blotting, an animal model of NAFLD was established by feeding SD rats a HFD for 60 days. Serum alanine aminotransferase (ALT), but not aspartate aminotransferase (AST), was significantly higher in HFD-fed than ND-fed rats (Supplementary Figure 1A, p < 0.05, and Supplementary Figure 1B). Both serum TG (Supplementary Figure 1C, p < 0.05) and intrahepatic TG (Supplementary Figure 1D, p < 0.01) were significantly higher in HFD-fed rats than in ND-fed rats. Representative photomicrographs of the liver were shown following HE staining of ND-fed (Fig. 1F) and HFD-fed (Fig. 1G) rats and IHC of SENP3 in livers from ND-fed (Fig. 1H) and HFD-fed (Fig. 1I) rats. The relative mean density of intrahepatic SENP3 was ~10 fold higher in HFD-fed rats compared with ND-fed rats (Fig. 1J, p < 0.01). Significant up-regulation of SENP3 in livers from HFD-fed rats compared with that from ND-fed rats (Fig. 1K) (Fig. 1L, p < 0.01), was confirmed using Western blotting.
Intrahepatic SENP3 production was determined in NAFLD and controls.
Histopathology of the liver tissues from healthy liver donors (n = 3) (A) and NAFLD (n = 8) (B) were H&E stained. Intrahepatic SENP3 production from healthy liver donors (C) and NAFLD (D) were labelled immunohistochemically, with the black arrows indicated the positive labelling. Quantification of immune-staining was presented (E, p < 0.001). The corresponding H&E of the liver from ND- (n = 5) (F) and HFD-fed (n = 5) (G) rats and immunohistochemistry of SENP3 in the liver from ND- (H) and HFD-fed (I) rats and their corresponding quantification were also presented (J, p < 0.001). The hepatic protein production of SENP3 from HFD-fed and ND-fed rats was confirmed by Western blotting (K) and the quantification was presented (L, p < 0.01).
Hepatic SENP3 in rats fed a HFD for 0, 30, 60 and 120 days was determined by Western blotting (Supplementary Figure 2A). SENP3 relative protein level was gradually up-regulated following HFD over 120 days, but appeared to plateau at day 60 (Supplementary Figure 2B). Furthermore, there was a significant correlation between hepatic SENP3 and TG (rho = 0.77, p < 0.001) (Supplementary Figure 2C). However, there was no significant correlation between hepatic SENP3 and severity of steatosis [(mild, 5–33%, n = 3) and (moderate, 33–66%, n = 5)] NAFLD patients (data not shown). The data in vivo above suggests a close relationship between SENP3 and NAFLD, which invites speculation that SENP3 is correlated with the development of NAFLD.
SENP3 contributed to the severity of hepatic steatosis in vitro
A model of hepatic steatosis was established in cultured hepatocytes to further explore the role of SENP3 in NAFLD in vitro. Compared with non-treated hepatocytes (Fig. 2A), intracellular lipid accumulation was substantially up-regulated in FFA-treated hepatocytes (Fig. 2B), using Oil Red O staining. Consistent with the in vivo findings above, significantly up-regulated SENP3 protein was detected in FFA-treated hepatocytes compared with non-treated hepatocytes in vitro, using Western blotting (Supplementary Figure 3A, B, p < 0.01). To verify if SENP3 contributes to the development of NAFLD, SENP3 was either silenced with SENP3-siRNA (Supplementary Figure 3C) or overexpressed with a GFP-SENP3 plasmid (Supplementary Figure 3D) in hepatocytes in vitro. Subsequently, these genetically manipulated cells were further treated with or without FFA for 12 hours. Compared with NS-siRNA (Fig. 2C), up-regulated lipid accumulation with FFA treatment was markedly ameliorated with SENP3-siRNA transfection (SENP3 inhibition) (Fig. 2D); whereas, compared with PCDNA3.1 (Fig. 2E), lipid accumulation was substantially increased with GFP-SENP3 transfection (SENP3 overexpression) (Fig. 2F). Consistent with our hypothesis, SENP3 silencing or overexpression resulted in a significant reduction (44%, p < 0.05) or up-regulation (~2 fold, p < 0.05) of cellular TG compared to NS-siRNA or PCDNA3.1 vehicle plasmid transfected group following FFA exposure (Fig. 2G). Collectively, these data suggest that SENP3 plays a critical role in steatosis.
SENP3 regulated lipid accumulation in hepatocytes.
Intrahepatic lipid was labelled with Oil red o in hepatocytes treated without FFA (A) and with FAA (B). Compared with NS-siRNA (C), FFA induced lipid accumulation was attenuated in hepatocyte transfected with SENP3-siRNA (D). Compared with PCDNA3.1 (E), FFA induced lipid content was further enhanced with GFP-SENP3 (F). Intrahepatic TG was quantified from different treatments (G). Intrahepatic SENP3 mRNA from the different treatments was quantified, using qRT-PCR (H,I).
qRT-PCR was applied to verify the expression of SENP3 in hepatocytes with different treatments. SENP3 was significantly up-regulated in GFP-SENP3 transfected hepatocytes (~3 fold, p < 0.05), whereas it was significantly down-regulated with SENP3-siRNA transfection (>50%, p < 0.05) (Fig. 2H). SENP3 mRNA level was up-regulated with FFA stimulation in mock-treated hepatocytes (p < 0.01), SENP3-siRNA transfected hepatocytes (p < 0.05) or GFP-SENP3 transfected hepatocytes (p < 0.01), compared with non-FAA-stimulated cells (Fig. 2I). As expected, we observed a 66% down-regulation in SENP3 in SENP3 silenced hepatocytes with FAA stimulation, compared with the non-treated hepatocytes with FFA stimulation (Fig. 2I, p < 0.01). On the other hand, 5 fold up-regulated SENP3 was detected in SENP3 overexpressed hepatocytes with FAA, compared with the non-treated hepatocytes with FFA stimulation (Fig. 2I, p < 0.01). Overall, these data provide further evidence that SENP3 plays an important role in steatosis.
Differentially expressed genes (DEGs) in NAFLD
To explore the role of SENP3 during the development of NAFLD, the expression of SENP3 and SENP3-related genes were determined during the development of NAFLD in vitro, using the RNA-Seq method. DEGs were detected using the GFOLD algorithm adjusting among the SENP3 silenced, overexpressed and the mock-gene treated groups following FFA stimulation. There were 140 DEGs identified in the SENP3 overexpressed hepatocytes following FFA treatment, compared to the mock-gene treated group, and 426 DEGs in SENP3 silenced hepatocytes following FFA treatment, compared to the mock-gene treated group. After combining these DEGs from the groups with overexpression and silencing of SENP3, 532 unique DEGs were generated. To determine which of these DEGs were of clinical value in NAFLD patients, potential secreted proteins among these DEGs were selected by searching against the ExoCarta diagnosis and prognosis database to find those likely to be secreted into plasma or serum24. It was identified that 91 out of 532 DEGs were potential secreted proteins (Supplementary Table 1). To enrich these DEGs functions, these 91 secreted genes were further submitted to the IPA pathway package against human species to accumulate the potential sub-network(s) or pathway(s) related to lipid metabolism (Supplementary Figure 4). Using NAFLD functional selection, it was further recognized that 11 out of 91 unique secreted genes were related to liver steatosis and steatohepatitis (Table 1). A sub-network enriched for lipid metabolic disorder was constructed based on these 11 recognized genes (Fig. 3). These secreted proteins might be potentially promising diagnostic biomarkers of NAFLD. The GFOLD value from the DEGs overexpression groups revealed the top 3 SENP3-related genes (apoe, a2m and tnfrsf11b), which were then utilized to confirm the clinical significance of SENP3 activity (Fig. 3).
Apoe, a2m and tnfrsf11b were regulated by SENP3 following FFA treatment in hepatocytes
Expression of apoe, a2m and tnfrsf11b was confirmed using qRT-PCR in hepatocytes. The apoe mRNA level was significantly inhibited (~60%, p < 0.001) in SENP3-siRNA transfected (SENP3 knocked-down) hepatocytes compared with NS-siRNA transfected hepatocytes (Fig. 4A). On the other hand, apoe was significantly up-regulated (>3 fold, p < 0.001) in GFP-SENP3 transfected (SENP3 overexpressed) hepatocytes compared with PCDNA3.1 transfected hepatocytes (Fig. 4A). Similarly, mRNA levels of a2m (Fig. 4B) and tnfrsf11b (Fig. 4C) were significantly inhibited with SENP3 silencing, or increased with SENP3 overexpression, consistent with the regulatory role of SENP3. There was also a significant up-regulation of apoe following FFA stimulation in mock-treated hepatocytes (p < 0.001), SENP3-silenced hepatocytes (p < 0.01) or SENP3-overexpressed hepatocytes (Fig. 4D, p < 0.05), consistent with data obtained from SENP3 expression above. Up-regulated apoe was detected in SENP3-overexpressed hepatocytes plus FAA stimulation, compared with mock-treated hepatocytes plus FFA stimulation (Fig. 4D, >2 fold, p < 0.01). As expected, ~70% down-regulation of apoe was detected in SENP3-silenced hepatocytes following FAA stimulation, compared with mock-treated hepatocytes plus FFA stimulation (Fig. 4D, p < 0.001). Interestingly, the constitutive expression of a2m (Fig. 4E) and tnfrsf11b (Fig. 4F) in hepatocytes was also significantly up-regulated with FFA stimulation. Moreover, a2m (Fig. 4E) and tnfrsf11b (Fig. 4F) were down-regulated in SENP3-silenced hepatocytes plus FFA, compared with mock-treated hepatocytes plus FFA, and up-regulated in SENP3-overexpressed hepatocytes plus FFA, compared with mock-treated hepatocytes plus FFA. The mRNA expression of a2m or tnfrsf11b modified by either SENP3 silencing or over expression with FFA was in line with the findings in apoe gene, but at different magnitudes. Taken together, mRNA levels of apoe, a2m and tnfrsf11b, regulated by SENP3, were up-regulated following FFA stimulation in vitro. Thus, these data are consistent with SENP3, contributing to hepatic steatosis via the 3 selected lipid metabolism-related genes.
Top three SENP3 related genes (apoe, a2m, and tnfrsf11b) were quantified in vitro using qRT-PCR. mRNA of apoe (A), a2m (B), and tnfrsf11b (C) in the hepatocytes was silenced with SENP3-siRNA transfection, but increased with GFP-SENP3 transfection. To determine the effects of SENP3 in lipid regulation, apoe (D), a2m (E) and tnfrsf11b (F) mRNA levels were further quantified in control, SENP3-siRNA transfected and GFP-SENP3 transfected hepatocytes with/without FFA stimulation.
Up-regulated intrahepatic and circulating APOE, A2M and TNFRSF11B in NAFLD patients
Following the data obtained in hepatocytes at mRNA level, the corresponding protein levels of apoe, a2m and tnfrsf11b were determined in the liver tissue from NAFLD patients and healthy liver transplant donors, as well as in the plasma from NAFLD patients and HCs. Constitutive protein production of APOE was observed in livers from healthy liver transplant donors (Fig. 5A, left) and NAFLD (Fig. 5A, right) patients. In NAFLD patients, APOE was mainly located in the cell membrane of hepatocytes and the adjacent extra-cellular space, particularly in regions containing many fat vacuoles. APOE was significantly increased (Fig. 5B, p < 0.001) in the hepatocytes from NAFLD patients, compared to those from the donors. A2M (Fig. 5C) and TNFRSF11B (Fig. 5E) in the liver from the donor (left panels) and NAFLD patients (right panels) were also detected immunohistochemically, with increased staining in NAFLD patients mainly located in the cytoplasm. A2M (Fig. 5D, p < 0.001) and TNFRSF11B (Fig. 5F, p < 0.001) were significantly up-regulated, compared with liver transplant donors. Thus, the expression of all three proteins was increased in liver tissue samples from NAFLD patients.
Intrahepatic and circulating APOE, A2M, and TNFRSF11B were determined in NAFLD patients and controls.
APOE (A), A2M (C), and TNFRSF11B (E) in the liver tissues from healthy liver donors (left panels, n = 3) and NAFLD (right panels, n = 8) were labelled immunohistochemically. The quantification of APOE (B, p < 0.001), A2M (D, p < 0.001), and TNFRSF11B (F, p < 0.001) were also presented. Circulating levels of APOE (G, p < 0.05), A2M (H, p < 0.05), and TNFRSF11B (I, p < 0.001) in NAFLD patients (n = 50) and HCs (n = 40) were verified by ELISA.
Liver biopsy is an invasive procedure during clinical practice, thus an alternative diagnostic approach is desirable to measure circulating APOE, A2M and TNFRSF11B. Hence, we used ELISA to measure circulating levels of these proteins. The circulating level of APOE was 2.5 fold higher in NAFLD patients than from HCs (Fig. 5G, p < 0.05). Similarly, circulating A2M (Fig. 5H, 1.38 fold, p < 0.05) and TNFRSF11B (Fig. 5I, 1.47 fold, p < 0.001) were significantly higher in NAFLD patients than HCs. These plasma data are consistent with the immunohistochemical findings in the liver tissues.
Discussion
In the current study we observed that SENP3 was increased in the liver from NAFLD patients and HFD-fed rats in vivo, as well as FFA-treated hepatocytes in vitro. Lipid accumulation in FFA-stimulated hepatocytes was inhibited with SENP3 silencing, but was enhanced with SENP3 overexpression. SENP3 related genes in NAFLD were generated using RNA-Seq from the hepatocytes in vitro. The top three genes (apoe, a2m and tnfrsf11b) were found to be regulated by SENP3 in hepatic steatosis in vitro. Furthermore, protein levels of APOE, A2M and TNFRSF11B were significantly up-regulated in the liver and plasma of NAFLD patients compared with the controls. Such data demonstrate a clear role for SENP3 in lipid metabolism during the development of NAFLD.
Mammals possess six SENP isoforms that are capable of reversing SUMO-1, -2 and -3 post-translational modifications. These isoforms play different roles in the control of various cellular events27. SENP2 regulates adipogenesis via de-SUMOylation of C/EBPβ, which in turn promotes C/EBPα and PPARγ28, and controls glucose metabolism via p-AKT9. Overexpression of SENP2 promotes fatty acid metabolism in skeletal muscle via fatty acid oxidation, ultimately alleviating obesity-linked metabolic disorders10. On the other hand, SENP2 silence attenuates adipogenesis in preadipocytes28. In addition, down-regulation of SENP2 increases SUMOylation of p53 and ERK5, leading to atherosclerotic plaque formation29. In the current study, hepatic production of SENP3 was significantly higher in NAFLD patients and HFD-fed rats than healthy liver transplant donors or ND-fed rats, as well as, in FFA-treated hepatocytes than non-treated cells.
A close correlation between hepatic TG and SENP3 in HFD fed rats suggests a possible role of SENP3 in lipid metabolism in NAFLD. However, the lack of a significant correlation between the severity of steatosis and hepatic SENP3 from NAFLD patients might be due to relatively small numbers of patients in this study and/or patient polymorphisms. There are both clinical and ethical challenges to performing liver biopsies in NAFLD patients. The large number of liver biopsies required for our future clarification of the correlation between the severity of steatosis and hepatic SENP3 are currently being collected.
The observation of the correlation between SENP3 and NAFLD in patients and the animal model in vivo was further clarified in hepatocytes in vitro. It was confirmed that increased lipid accumulation in FFA-treated hepatocytes was attenuated or enhanced with SENP3 silencing or overexpression, respectively, arguing for the regulation of hepatic steatosis by SENP3. The corresponding mRNA levels of SENP3 were also confirmed. Thus, the data delineated that SENP3 activity contributes to the severity of steatosis, suggesting a potential novel therapeutic target for the management of hepatic steatosis. We observed that SENP3 is up-regulated in livers from NAFLD patients and HFD fed rats in vivo, and after loading hepatocytes with free fatty acids (FFA) in vitro. We also observed that SENP3 overexpression resulted in a significant up-regulation of cellular TG compared with control group following FFA exposure. Thus, we believe that SENP3 likely acts in both paracrine and autocrine fashions during steatosis. However, the precise underlying mechanism of SENP3 in steatosis will be determined in future experiments, using SENP3 liver-specific knockout mice.
Subsequently, the differential expression of 11 SENP3-related genes were identified in the NAFLD disease state, following functional selection from 91 secreted genes out of 532 DEGs. Three out of these 11 genes (apoe, a2m and tnfrsf11b) were subsequently selected to validate their significance in NAFLD. Expression of apoe, a2m and tnfrsf11b genes were elevated in steatotic hepatocytes and were regulated by SENP3 at the mRNA level. APOE, a ligand for lipoprotein receptors, regulates lipid metabolism via participating in lipid transportation and promoting lipid accumulation30. The critical role of APOE in metabolic syndrome has been well documented in APOE gene knock-out mice31. Interestingly, APOE, regulated by SENP3, was elevated in steatotic hepatocytes, as well as, in the liver and plasma from the NAFLD patients. There is a close correlation between elevated APOE and hyperlipidemia in metabolic syndrome patients32. We speculate that the elevated APOE in NAFLD may be regulated by SENP3 indirectly. The up-regulation of APOE may be a response by the body to metabolize the elevated lipid levels in the liver of the NAFLD patients we examined. The APOE response in NAFLD may be functionally insufficient or inefficient, which is being currently investigated.
A2M can promote cell proliferation by increasing glucose uptake, lactate secretion and lipogenesis, as an insulin-like response33. Abnormal up-regulation of A2M is associated with intracellular lipid accumulation in a new model of NAFLD34. Such findings are consistent with the data obtained in our current study, i.e. the observation of significantly elevated circulating and intrahepatic A2M in NAFLD patients, as well as, in steatotic hepatocytes in vitro. Our data imply that A2M, regulated by SENP3, plays an important role in the progression of NAFLD.
TNFRSF11B (Osteoprotegerin, OPG) is a secreted protein involved in bone turnover, due to its role as a decoy receptor for the receptor activator of nuclear factor-kB ligand in the osteoclasts35. It has been reported that there is a linkage between OPG and NAFLD, particularly in osteoporosis patients36, suggesting a possible role of OPG in the development of NAFLD. Our current study showed that OPG was substantially up-regulated at the mRNA level in steatotic hepatocytes in vitro and at the protein level in NAFLD patients. Our data suggest that OPG contributes to lipid metabolism in NAFLD, which is in line previous findings36.
In addition to apoe, a2m and tnfrsf11b, substantial alterations in the expression of several other genes were identified during our sub-network analysis (Fig. 3). Relevant examples are as follows. ATP-binding cassette transporter 1 (abcb1), which participates in drug metabolism, is a monitor for the efficacy and safety of treatment of NAFLD37,38. Caspase-9 (casp9), is a potential indicator for hepatocyte apoptosis during the development of NAFLD via the mitochondrial pathway39. FABP3 is a regulator of lipid metabolism and participates in the transport of lipids40, and has been used as a biomarker for metabolic syndrome related atherosclerosis in patients with glucose impairment41. Serpine 1 (plasminogen activator inhibitor-1), which correlates to body-mass index, plasma triglyceride and insulin levels, has a significant clinical value in metabolic disturbance related diseases, including atherosclerosis, type 2 diabetes, obesity and liver steatosis42,43. Our findings suggest that SENP3 plays an important role in homeostasis of lipid metabolism, and dysregulation of SENP3 contributes to consequent NAFLD via gene regulation e.g. apoe, a2m and tnfrsf11b.
There are limitations in the current study. Firstly, the direct linkage between SENP3 and NAFLD has not been fully established in a SENP3 knockout animal model, due to embryonic lethality. However, such a lethal consequence further supports the critical pathophysiological role of SENP3 in homeostasis. Secondly, a larger patient cohort, including population diversity, is desirable for the validation of the 11 selected secreted genes with potential diagnostic values in NAFLD. Thirdly, the precise underlying mechanism of SENP3 regulation during the development of NAFLD is currently being investigated.
The focus of the current study was to investigate whether SENP3 contributes to lipid metabolism in NAFLD, based on our unexpected preliminary observation that SENP3 is up-regulated in fatty liver. Subsequently, follow up in vivo and in vitro experiments demonstrated a link between SENP3 and NAFLD. There are a number of targets of SENP3 e.g. MEF2D, EP300, RbBP5, NPM1, etc, involved in transcriptional activation, potentiating cell survival and redox regulation44. Three molecules (apoe, a2m and tnfrsf11b) chosen in the current study were based on the data from bioinformatics, which offered the highest probability of a linkage between SENP3 and these three molecules. We also observed differential regulation of apoe, a2m and tnfrsf11b following manipulation of SENP3. We acknowledge that the underlying mechanism is still unclear. Nevertheless, our current observations suggest that SENP3 regulation of lipid metabolism in fatty liver may possibly be via apoe, a2m and tnfrsf11b. In addition, it has been reported that SENP3 serves as a redox sensor to enhance HIF1A transcriptional activity by de-SUMOylating EP300 45,46. We will use liver-specific conditional SENP3 knockout mice to investigate the precise role of SENP3 in future experiments.
In conclusion, we observed for the first time that SENP3 was up-regulated in NAFLD patients and an animal model in vivo, as well as steatotic hepatocytes in vitro. Moreover, SENP3-mediated steatosis occurs possibly via regulating downstream genes involved in abnormal lipid metabolism. Such data might shed light on the pathogenesis of steatosis, which may be used as a potential therapeutic target for prevention and/or treatment of NAFLD. The precise role of SENP3 in the development of NAFLD will continue to be studied.
Additional Information
How to cite this article: Liu, Y. et al. SUMO-specific protease 3 is a key regulator for hepatic lipid metabolism in non-alcoholic fatty liver disease. Sci. Rep. 6, 37351; doi: 10.1038/srep37351 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Rinella, M. E. Nonalcoholic fatty liver disease: a systematic review. Jama 313, 2263–2273, doi: 10.1001/jama.2015.5370 (2015).
Musso, G., Gambino, R. & Cassader, M. Recent insights into hepatic lipid metabolism in non-alcoholic fatty liver disease (NAFLD). Prog Lipid Res 48, 1–26, doi: 10.1016/j.plipres.2008.08.001 (2009).
Kawano, Y. & Cohen, D. E. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol 48, 434–441, doi: 10.1007/s00535-013-0758-5 (2013).
Sahini, N. & Borlak, J. Recent insights into the molecular pathophysiology of lipid droplet formation in hepatocytes. Prog Lipid Res 54, 86–112, doi: 10.1016/j.plipres.2014.02.002 (2014).
Chalasani, N. et al. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 55, 2005–2023, doi: 10.1002/hep.25762 (2012).
Yeh, E. T. SUMOylation and De-SUMOylation: wrestling with life’s processes. J Biol Chem 284, 8223–8227, doi: 10.1074/jbc.R800050200 (2009).
Nayak, A. & Muller, S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol 15, doi: Artn 422 10.1186/S13059-014-0422-2 (2014).
Cai, R. et al. SUMO-specific protease 1 regulates mitochondrial biogenesis through PGC-1alpha. J Biol Chem 287, 44464–44470, doi: 10.1074/jbc.M112.422626 (2012).
Tang, S. et al. Role of SUMO-specific protease 2 in reprogramming cellular glucose metabolism. PloS one 8, e63965, doi: 10.1371/journal.pone.0063965 (2013).
Koo, Y. D. et al. SUMO-Specific Protease 2 (SENP2) Is an Important Regulator of Fatty Acid Metabolism in Skeletal Muscle. Diabetes 64, 2420–2431, doi: 10.2337/db15-0115 (2015).
Zunino, R., Schauss, A., Rippstein, P., Andrade-Navarro, M. & McBride, H. M. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J Cell Sci 120, 1178–1188, doi: 10.1242/jcs.03418 (2007).
Gong, L. & Yeh, E. T. H. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem 281, 15869–15877, doi: 10.1074/jbc.M511658200 (2006).
Han, Y. et al. SENP3-mediated De-conjugation of SUMO2/3 from Promyelocytic Leukemia Is Correlated with Accelerated Cell Proliferation under Mild Oxidative Stress. J Biol Chem 285, 12906–12915, doi: 10.1074/jbc.M109.071431 (2010).
Huang, C. et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. Embo J 28, 2748–2762, doi: 10.1038/emboj.2009.210 (2009).
Wei, H. X. et al. An Upregulation of SENP3 After Spinal Cord Injury: Implications for Neuronal Apoptosis. Neurochem Res 37, 2758–2766, doi: 10.1007/s11064-012-0869-z (2012).
Guo, C. et al. SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia. Embo J 32, 1514–1528, doi: 10.1038/emboj.2013.65 (2013).
Ren, Y. H. et al. De-SUMOylation of FOXC2 by SENP3 promotes the epithelial-mesenchymal transition in gastric cancer cells. Oncotarget 5, 7093–7104 (2014).
Lieber, C. S. et al. Model of nonalcoholic steatohepatitis. The American journal of clinical nutrition 79, 502–509 (2004).
Wang, M. et al. SENP3 regulates the global protein turnover and the Sp1 level via antagonizing SUMO2/3-targeted ubiquitination and degradation. Protein & cell, doi: 10.1007/s13238-015-0216-7 (2015).
Joshi-Barve, S. et al. Palmitic acid induces production of proinflammatory cytokine interleukin-8 from hepatocytes. Hepatology 46, 823–830, doi: 10.1002/hep.21752 (2007).
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967, doi: 10.1093/bioinformatics/btp336 (2009).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods 5, 621–628, doi: 10.1038/nmeth.1226 (2008).
Feng, J. et al. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 28, 2782–2788, doi: 10.1093/bioinformatics/bts515 (2012).
Keerthikumar, S. et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J Mol Biol., doi: 10.1016/j.jmb.2015.09.019 (2015).
An, F. et al. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis: an international journal on programmed cell death 17, 702–716, doi: 10.1007/s10495-012-0704-7 (2012).
Xiang, X. et al. IL-22 and non-ELR-CXC chemokine expression in chronic hepatitis B virus-infected liver. Immunology and cell biology 90, 611–619, doi: 10.1038/icb.2011.79 (2012).
Hickey, C. M., Wilson, N. R. & Hochstrasser, M. Function and regulation of SUMO proteases. Nature reviews. Molecular cell biology 13, 755–766, doi: 10.1038/nrm3478 (2012).
Chung, S. S. et al. Control of adipogenesis by the SUMO-specific protease SENP2. Mol Cell Biol 30, 2135–2146, doi: 10.1128/MCB.00852-09 (2010).
Heo, K. S. et al. De-SUMOylation enzyme of sentrin/SUMO-specific protease 2 regulates disturbed flow-induced SUMOylation of ERK5 and p53 that leads to endothelial dysfunction and atherosclerosis. Circulation research 112, 911–923, doi: 10.1161/CIRCRESAHA.111.300179 (2013).
Mahley, R. W. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science 240, 622–630 (1988).
Hofmann, S. M. et al. Defective lipid delivery modulates glucose tolerance and metabolic response to diet in apolipoprotein E-deficient mice. Diabetes 57, 5–12, doi: 10.2337/db07-0403 (2008).
Onat, A. et al. High serum apolipoprotein E determines hypertriglyceridemic dyslipidemias, coronary disease and apoA-I dysfunctionality. Lipids 48, 51–61, doi: 10.1007/s11745-012-3724-8 (2013).
Misra, U. K. & Pizzo, S. V. Activated alpha(2)-Macroglobulin Binding to Human Prostate Cancer Cells Triggers Insulin-like Responses. J Biol Chem 290, 9571–9587, doi: 10.1074/jbc.M114.617837 (2015).
Chavez-Tapia, N. C., Rosso, N. & Tiribelli, C. Effect of intracellular lipid accumulation in a new model of non-alcoholic fatty liver disease. Bmc Gastroenterol 12, doi: Artn 20 10.1186/1471-230x-12-20 (2012).
Baud’huin, M. et al. Osteoprotegerin: multiple partners for multiple functions. Cytokine Growth Factor Rev 24, 401–409, doi: 10.1016/j.cytogfr.2013.06.001 (2013).
Yilmaz, Y. Review article: non-alcoholic fatty liver disease and osteoporosis–clinical and molecular crosstalk. Alimentary pharmacology & therapeutics 36, 345–352, doi: 10.1111/j.1365-2036.2012.05196.x (2012).
Hardwick, R. N., Fisher, C. D., Street, S. M., Canet, M. J. & Cherrington, N. J. Molecular Mechanism of Altered Ezetimibe Disposition in Nonalcoholic Steatohepatitis. Drug Metab Dispos 40, 450–460, doi: 10.1124/dmd.111.041095 (2012).
Hardwick, R. N. et al. Increased Susceptibility to Methotrexate-Induced Toxicity in Nonalcoholic Steatohepatitis. Toxicol Sci 142, 45–55, doi: 10.1093/toxsci/kfu156 (2014).
Wurstle, M. L., Laussmann, M. A. & Rehm, M. The central role of initiator caspase-9 in apoptosis signal transduction and the regulation of its activation and activity on the apoptosome. Exp Cell Res 318, 1213–1220, doi: 10.1016/j.yexcr.2012.02.013 (2012).
Chmurzynska, A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. Journal of applied genetics 47, 39–48, doi: 10.1007/BF03194597 (2006).
Karbek, B. et al. Heart-Type Fatty Acid Binding Protein (H-FABP): Relationship with arterial intima-media thickness and role as diagnostic marker for atherosclerosis in patients with impaired glucose metabolism. Cardiovasc Diabetol 10, doi: Artn 37 10.1186/1475-2840-10-37 (2011).
Iwaki, T., Urano, T. & Umemura, K. PAI-1, progress in understanding the clinical problem and its aetiology. British journal of haematology 157, 291–298, doi: 10.1111/j.1365-2141.2012.09074.x (2012).
Barbato, A. et al. Relationships of PAI-1 levels to central obesity and liver steatosis in a sample of adult male population in southern Italy. Internal and emergency medicine 4, 315–323, doi: 10.1007/s11739-009-0240-9 (2009).
Huang, C. J., Wu, D., Khan, F. A. & Huo, L. J. DeSUMOylation: An Important Therapeutic Target and Protein Regulatory Event. DNA and cell biology 34, 652–660, doi: 10.1089/dna.2015.2933 (2015).
Wang, Y. et al. The biphasic redox sensing of SENP3 accounts for the HIF-1 transcriptional activity shift by oxidative stress. Acta Pharmacol Sin 33, 953–963, doi: 10.1038/Aps.2012.40 (2012).
Huang, C. et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. Embo J 28, 2748–2762, doi: 10.1038/emboj.2009.210 (2009).
Acknowledgements
We appreciate Prof Jinke Cheng for constructive comments. This work was supported by the Shanghai Leading Talent Program, the National Natural Science Foundation of China (81171569, 81570535), Shanghai Science and Technology Committee (134119a2400, 12XD1403600), Doctoral Innovation Fund Projects from Shanghai Jiaotong University School of Medicine (BXJ201415), the National Clinical Key Specialty Construction Project of China (Infectious Diseases), Fourth key subject construction project of Shanghai Public Health three year plan of action (15GWZK0102) and the Shanghai Three-Year Plan of the Key Subjects Construction in Public Health-Infectious Diseases and Pathogenic Microorganism (15GWZK0102).
Author information
Authors and Affiliations
Contributions
Study concept and design: Y.H.L. and Q.X.; data collection: Y.H.L., Y.H., L.Q., Z.J.C., X.G.X., S.W.J., X.L.W., J.L., R.T.L; data interpretation: W.C., H.W., S.S.B., Q.X., L.Y.H. and F.D.Y.; drafting the manuscript: L.Y.H. and F.D.Y.; critical revision of the manuscript: S.S.B. and Q.X. All authors had access to the study data and have reviewed and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Liu, Y., Yu, F., Han, Y. et al. SUMO-specific protease 3 is a key regulator for hepatic lipid metabolism in non-alcoholic fatty liver disease. Sci Rep 6, 37351 (2016). https://doi.org/10.1038/srep37351
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep37351
This article is cited by
-
SENP1 prevents steatohepatitis by suppressing RIPK1-driven apoptosis and inflammation
Nature Communications (2022)
-
Correlation between hepatic human males absent on the first (hMOF) and viral persistence in chronic hepatitis B patients
Cell & Bioscience (2018)
-
Berberine ameliorates blockade of autophagic flux in the liver by regulating cholesterol metabolism and inhibiting COX2-prostaglandin synthesis
Cell Death & Disease (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.