Abstract
Viroids are plant-pathogenic molecules made up of single-stranded circular non-coding RNAs. How replicating viroids interfere with host silencing remains largely unknown. In this study, we investigated the effects of a nuclear-replicating Potato spindle tuber viroid (PSTVd) on interference with plant RNA silencing. Using transient induction of silencing in GFP transgenic Nicotiana benthamiana plants (line 16c), we found that PSTVd replication accelerated GFP silencing and increased Virp1 mRNA, which encodes bromodomain-containing viroid-binding protein 1 and is required for PSTVd replication. DNA methylation was increased in the GFP transgene promoter of PSTVd-replicating plants, indicating involvement of transcriptional gene silencing. Consistently, accelerated GFP silencing and increased DNA methylation in the of GFP transgene promoter were detected in plants transiently expressing Virp1. Virp1 mRNA was also increased upon PSTVd infection in natural host potato plants. Reduced transcript levels of certain endogenous genes were also consistent with increases in DNA methylation in related gene promoters in PSTVd-infected potato plants. Together, our data demonstrate that PSTVd replication interferes with the nuclear silencing pathway in that host plant, and this is at least partially attributable to Virp1. This study provides new insights into the plant-viroid interaction on viroid pathogenicity by subverting the plant cell silencing machinery.
Similar content being viewed by others
Introduction
Many eukaryotes share highly abundant classes of small RNAs (sRNAs) that range in size from 20 to 25 nucleotides (nt) produced from longer double-stranded RNA (dsRNA) precursors through the action of RNase III-like Dicer or Dicer-like (DCL) enzymes1,2. The sRNAs participate in RNA silencing (or RNA interference, RNAi) by binding to an ARGONAUTE (AGO) protein in RNA-induced silencing complexes (RISCs) and act as sequence-specificity determinants through RNA-RNA and also RNA-DNA interactions. These interactions mediate a wide range of phenomena such as post-transcriptional gene silencing (PTGS) of mRNA and transcriptional silencing (TGS) of heterochromatin, as well as translational inhibition of mRNA3,4,5. In plants, the TGS pathway consists of RNA-directed DNA methylation (RdDM), which requires both 24-nt small interfering RNAs (siRNAs) and long non-coding RNA transcripts as well as proteins, such as DNA-dependent RNA polymerase IV (Pol IV), RNA-dependent RNA polymerase 2 (RDR2) and AGO4 for de novo DNA methylation. This pathway has a great impact on the expression and constitution of the plant genome5.
In addition to regulation of genome integrity and development, RNAi has roles in transposon restriction and anti-pathogen defense1,6,7,8,9. As a counterstrategy to host RNA silencing, many viruses encode suppressor proteins (VSR) targeting DCL, RISC or small RNA activities10,11,12,13,14. sRNAs derived from some subviruses, such as viroids and viral satellites, were also detected in infected plants, suggesting that these unconventional RNAs are also targeted by PTGS15,16,17,18,19,20,21,22,23,24. However, these subviruses may possess novel silencing suppressor activities that counter-defend against host silencing. The function of these subviruese is attributed to the resistance of their secondary structures to RISC-mediated cleavage rather than directly targeting the host silencing components as some VSRs do25,26.
Viroids are the smallest of transmissible molecules, ranging from 246 to 401 nucleotides in size27, and are made up of single-stranded circular RNA that is non-translatable and autonomously replicates by redirected host processes27,28. A type member of the Pospiviroidae viroid family, Potato spindle tuber viroid (PSTVd) replicates in the nucleus via an asymmetric rolling circle mechanism with the aid of host-encoded DNA-dependent RNA polymerase II28,29,30,31. A host RNA-binding protein, viroid-binding protein 1 (Virp1), was shown to interact with PSTVd and was required for PSTVd infection in both Nicotiana plants and protoplasts32,33. Early work by Wassenegger and colleagues on viroid-host interactions led to the landmark discovery of RNA-directed DNA methylation (RdDM). These researchers introduced PSTVd cDNA into the tobacco genome and found that the autonomous viroid RNA-RNA replication triggered viroid cDNA methylation34.
It has recently been reported that infection of cucumbers with a nuclear-replicating Hop stunt viroid (HSVd) is accompanied by changes in DNA methylation of host ribosomal RNA genes35. However, the molecular basis of nuclear-replicating viroid interference with host silencing pathways remains largely unknown. In this study, we investigated the effects of PSTVd replication on induction of a transgene GFP silencing in N. benthamiana and found that PSTVd replication accelerated, rather than suppressed, GFP silencing in the transient expression system in the 16c GFP plant. Consistently, DNA methylation was increased in the GFP transgene promoter of PSTVd-infected 16c plants and was associated with Virp1, whose coding mRNA was induced by PSTVd replication. We also found that Virp1 mRNA was increased upon PSTVd infection in natural host potato plants. Reduced transcription of certain endogenous genes was also consistent with the increases in DNA methylation in related gene promoters in PSTVd-infected plants in which accumulation of tested endogenous miRNAs was not affected. Together, our results demonstrate that the host Viroid-binding bromodomain-containing protein, Virp1, which is required and induced by PSTVd infection, may partially contribute to the interference of the nuclear silencing pathway in PSTVd-infected plants.
Materials and Methods
Plant Materials and Growth Conditions
The GFP-transgenic Nicotiana benthamiana (line 16C) was described in previous study13. In this system, an Agrobacterium strain expressing 35S-GFP that is infiltrated to 16c plants induced silencing of the GFP transgene, resulting in red fluorescence. The cultivar Solanum tuberosum (KEXIN NO. 18) and Nicotiana benthamiana plants were grown in a glass house at 25 °C with16-h-light/8-h-dark cycles.
Cloning and Constructs
For the 35S-PSTVd construct, full-length PSTVd cDNAs were amplified by RT-PCR using total RNAs extracted from PSTVd infected potato leaves as the template. The RT-PCR products were cloned into T-Vectors (Tsingke) using T4 DNA Ligase (Thermo). After confirmation by sequencing, two full-length minus strand PSTVd cDNAs were tandemly linked and subcloned into the pCAMBIA1300-221 binary vectors. For 35S-∆PSTVd construct, the 340-bp full-length minus strand PSTVd RT-PCR product in T-Vectors was subcloned into the pCAMBIA1300-221 binary vectors. Replicative 35S-PSTVd and non-replicative 35S-∆PSTVd were confirmed using transient expression system in N. benthamiana and RNA gel blotting analysis (Supplementary Figure S1).
For the 35S-Virp1 construct, the 1848-bp sequence of the Virp1 gene was amplified from N. benthamiana, and the RT-PCR products were cloned into T-Vectors (Tsingke) using T4 DNA Ligase (Thermo). After confirmation by sequencing, NbVirp1 sequence was subcloned into the pCAMBIA1300-221 binary vectors. The NbVirp1 and Virp1 from Solanum tuberosum (StVirp1) sequences share 87% identity at the nucleotide level (Supplementary Figure S2).
For the Virp1i construct, the 158bp sense and antisense sequences of the middle part of the bromodomain of NbVirp1 were amplified, and the two PCR fragments were each cloned into T-vectors (Tsingke) using T4 DNA Ligase (Thermo). After confirmation by sequencing, the two sequences were inserted into an intron-containing intermediate construct (pSK-int)36, to obtain sequence cassettes containing the inverted-repeat RNAi constructs as previously described36, producing pSK-Virp1i. The cassettes were then subcloned into the pCAMBIA1300-221 binary vector to generate Virp1i.
A diagram of all constructs used in this study is shown in Supplementary Figure S3.
Viroid Inoculation and Transient Expression
Two-week-old Solanum tuberosum were inoculated with sap prepared freshly from wild-type PSTVd infected plants by rubbing mechanically. The phosphate buffer inoculated leaves were used as mock.
For transient assays, the EHA105 strain of Agrobacterium tumefaciens was transformed with the 35S-GFP, 35S-PSTVd, 35S-∆PSTVd, 35S-Virp1, 35S-Virp1i, 35S-GUS or 35S-P19 by electroporation, and transformants were selected on Luria-Bertani medium containing rifampicin at 10 mg/liter and kanamycin at 50 mg/liter. Equal volumes of Agrobacterium cultures containing 35S-GFP (OD600 1.0) with either 35S-PSTVd, 35S-∆PSTVd, 35S-Virp1 or 35S-P19, 35S-GUS control (OD600 1.5) were mixed and co-infiltrated into 5-week-old 16c leaves. For Virp1i infiltration, Agrobacterium cultures containing Virp1i (OD600 1.0) were infiltrated into 16c leaves. The endogenous NbVirp1 was silenced at 3dpi, and the silenced leaves were then used for the co-infiltration assay.
RNA Extraction and RNA Gel Blot Analysis
Total RNA was extracted from Agrobacterium-infiltrated N. benthamiana or viroid inoculated S. tuberosum leaf tissues using the hot-phenol method as previously described13. For high molecular weight RNA gel blots, 15 μg of total RNA was separated on 1.2% agarose gels containing 6% formaldehyde and transferred to nylon N+ membrane. For mature PSTVd and siPSTVd detection, the [a-32P]-UTP-labelled T7 RNA transcripts from cDNA clones in pGEM-Teasy vector using the MAXIscriptkit (Ambion) were used as probes; for GFP mRNA detection, the full length GFP were radioactively labelled by [a-32P]-dCTP using a Rediprime™II Random Prime Labelling System(GE healthcare), for siGFP detection, the [a-32P]-UTP-labelled T7 RNA transcripts from pGEM-Teasy-GFP vector using the MAXIscriptkit (Ambion) were used as probes; for Virp1 mRNA detection, the full length NbVirp1 were radioactively labelled by [a-32P]-dCTP using a Rediprime™II Random Prime Labelling System(GE healthcare); for miR167, miR159 and U6 detection, [r-32P]ATP-labeled specific oligonucleotide probe sequences were used.
Dot-blot hybridization detection of PSTVd accumulation was performed as described (Monger WA et al. 2015). The PSTVd DNA fragment was labeled using the DIG DNA PCR labeling kit (Mylab Corporation).
DNA Bisulfite Sequencing Analysis
DNA bisulfite sequencing analysis: incubation of the target DNA with sodium bisulfite results in conversion of unmethylated cytosine residues into uracil, leaving the methylated cytosines unchanged. Total DNA was extracted using the DNeasy Plant Mini Kit (Qiagen). A total of 1μg of DNA was used for bisulfate treatment using the EpiTect Bisulfite kit (Qiagen). The purified bisulfite-treated DNA was amplified, ligated into the pGEM-T vector (Tsingke), and sequenced. The cytosine methylation analysis was performed via the CyMATE program (http://www.cymate.org/).
Quantitative RT-PCR
Total RNA was extracted from Solanum tuberosum leaves using TRIzol reagent (Thermo). Genomic DNA was digested using DNase I (Takara) and then reverse-transcribed into cDNA using the GoScript™ Reverse Transcription System (Promega). qRT-PCR analysis was performed with a 1000 series Thermal Cycling Platform (Bio-Rad) using EvaGreen 2X qPCR MasterMIX (Applied Biological Materials Inc.). At least three biological replicates and three technical replicates within an experiment for each sample were performed.
McrBC Digestion-PCR
McrBC is an endonuclease which cleaves DNA containing methylcytosine on one or both strands, McrBC will not act upon unmethylated DNA. S. tuberosum genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen). A total of 3 μg of genomic DNA was digested by McrBC (Biolabs) at 37 °C overnight, precipitated by ethanol, and dissolved with nuclease-free water. A total of 40 ng McrBC-digested DNA was used for each PCR reaction.
All primer and probe sequences are listed in Supplementary Table S1.
Results
PSTVd accelerated GFP silencing in transient assays
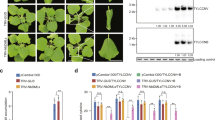
To investigate the effects of PSTVd replication on interference with plant RNA silencing processes, we first created a construct consisting of two tandem linked full-length minus strand PSTVd cDNAs under the control of the 35S promoter, resulting in 35S-PSTVd. Autonomous replication of the PSTVd RNA was confirmed by detection of mature PSTVd RNA in transient expression of 35S-PSTVd in the leaves of 16c using an Agro-infiltration assay (Fig. 1A). To test the effect on RNA silencing, Agrobaterium strains expressing 35S-PSTVd and 35S-GFP were co-infiltrated into the leaves of 16c plants. To our surprise, co-infiltration with 35S-PSTVd resulted in a significant low-intensity GFP green fluorescence in the infiltrated areas (Fig. 1B), compared to co-infiltration of 35S-GFP with the 35S-GUS control in the same leaves at 2 days post-infiltration (dpi) (Fig. 1B), where the GFP green fluorescence was maintained up to 3 dpi and began to decrease at 4 dpi (Fig. 1B). Red fluorescence appeared in areas co-infiltrated with 35S-GFP/35S-PSTVd, indicating silencing of the GFP transgene (Fig. 1B).
Effects of PSTVd replication on GFP silencing in transient assays.
(A) RNA gel blot detection of mature PSTVd RNA, GFP mRNA, PSTVd-derived siRNA (siPSTVd) and GFP-derived siRNA (siGFP) accumulation in 16c plants co-infiltrated with 35S-GFP and either 35S-GUS control or 35S-PSTVd at 4 dpi. Methylene blue-stained ribosome rRNA and U6 hybridization were used as the loading control. (B) Observation of local GFP silencing in the leaves of 16c plants co-infiltrated with 35S-GFP and either 35S-GUS control or 35S-PSTVd. Photographs were taken under UV light at 2, 3 and 4 dpi. (C) Observation of systemic GFP silencing spread to the whole plant induced by co-infiltrated 35S-GFP with either 35S-GUS control or 35S-PSTVd in separate plants. Photographs were taken under UV light at 4, 5 and 8 dpi.
To better observe GFP systemic silencing, we co-infiltrated 35S-GFP with 35S-PSTVd or 35S-GUS in separate plants. All plants co-infiltrated with 35S-PSTVd displayed systemic GFP silencing at 3 dpi and almost complete silencing at 5 dpi (Fig. 1C). At this point in plants co-infiltrated with 35S-GFP/35S-GUS, GFP silencing was only found in some infiltrated areas (Fig. 1C at 4 dpi), while systemic silencing occurred at 6–8 dpi (Fig. 1C). Similar results were observed in three independent experiments in which a total of 36 plants were used for each infiltration combination. Observation of both local and systemic GFP silencing suggested that co-infiltration with 35S-PSTVd might accelerate rather than suppress GFP silencing in the transient assay system.
Infiltrated leaves at 4 dpi were collected for total RNA and small RNA extraction for RNA gel blotting analysis. Large amounts of the siRNA derived from PSTVd (siPSTVd) were detected in leaves co-infiltrated with 35S-PSTVd (Fig. 1A), consistent with previous reports that viroid RNAs can trigger RNA silencing15,26. The GFP mRNA in leaves co-infiltrated with 35S-PSTVd accumulated at lower levels than those co-infiltrated with 35S-GUS, consistent with the low-intensity GFP fluorescence observed in 35S-PSTVd co-infiltrated leaves. However, the small RNA of GFP (siGFP) was detected with a significantly lower accumulation level in leaves co-infiltrated with 35S-PSTVd compared to leaves co-infiltrated with 35S-GUS (Fig. 1A). This finding contradicted the prevalent view that lower accumulation of GFP mRNA is normally accompanied by higher GFP-derived siRNA accumulation in the transient assay. Our result suggested that the low accumulation level of GFP mRNA did not entirely result from PTGS-mediated degradation of GFP mRNA that produced siGFP and implied that nuclear-replicating PSTVd may interfere with the expression of GFP mRNA at the transcriptional level.
PSTVd-induced host viroid-binding protein played a role in the acceleration of GFP silencing
To detect whether the 35S promoter-derived replication of the PSTVd sequence or the replicating PSTVd viroid caused the enhancement of GFP silencing, a non-replicative full-length minus strand PSTVd cDNA under the 35S promoter was constructed, resulting in 35S-ΔPSTVd. 16c plants were co-infiltrated with 35S-GFP and35S-GUS, 35S-PSTVd or 35S-ΔPSTVd. An intense GFP green fluorescence area was observed after 35S-GFP co-infiltration with 35S-GUS or 35S-∆PSTVd compared to areas co-infiltrated with 35S-PSTVd (Fig. 2A), indicating that replication of PSTVd was required for the enhancement of GFP silencing. Previous studies showed that PSTVd interacts with the host RNA-binding protein, Virp132,33. We therefore examined the response of Virp1 upon PSTVd replication in 16c plants. Increased accumulation of Virp1 mRNA was detected when 35S-PSTVd was infiltrated into 16c plants leaves compared to non-infiltrated 16c leaves with or without 35S-∆PSTVd infiltration (Fig. 2B). Replication of PSTVd was confirmed in the 35S-PSTVd-infiltrated leaves (Fig. 2B). Silencing of Virp1 in 16c leaves by infiltration of an RNAi construct, Virp1i, restricted PSTVd replication (Fig. 2B), which was consistent with the early finding that Virp1 was required for PSTVd infection in Nicotiana plants33. We then examined the effect of Virp1 on the PSTVd-accelerated GFP silencing process. Co-infiltration of 35S-GFP with 35S-PSTVd or 35S-Virp1 was then performed in 16c plants. Co-infiltration of 35S-GFP with 35S-GUS or 35S-P19, a viral suppressor protein, was used the control. The GFP green fluorescence rapidly decreased by 4 dpi after co-infiltration with 35S-PSTVd or 35S-Virp1, whereas green fluorescence was maintained after co-infiltration with controls at this time point (Fig. 2C). This result demonstrates that the reduced GFP mRNA level (Fig. 1A) is not attributable to replicative competition by the propagative PSTVd and also suggests that Virp1, which was induced by propagative PSTVd played at least a partial role in this enhanced silencing process.
Effects of PSTVd-induced host Virp1 on the acceleration of GFP silencing in transient assays.
(A) GFP fluorescence in the leaves of 16c plants co-infiltrated with 35S-GFP and either 35S- GUS control or 35S-PSTVd or 35S-∆PSTVd. Photographs were taken under UV light at 2 dpi. (B) RNA gel blot detection of Virp1 mRNA and mature PSTVd RNA accumulation in 16c plants and Virp1 silencing leaves co-infiltrated with 35S-GFP and either 35S-GUS control or 35S-PSTVd or 35S-∆PSTVd at 4 dpi. Methylene blue-stained ribosome rRNA was used as the loading control. (C) GFP fluorescence in leaves of 16c plants co-infiltrated with 35S-GFP and either 35S-GUS, 35S-P19 control or 35S-PSTVd or 35S-Virp1. Photographs were taken under UV light at 4 dpi.
PSTVd replication, Virp1 reduced expression and increased DNA methylation of the GFP transgene
We then examined the accumulation level of GFP mRNA after co-infiltration of 35S-GFP with 35S-Virp1. Total RNA was isolated from co-infiltrated leaves of the 16c plant. The accumulation of GFP mRNA was reduced in leaves co-infiltrated with 35S-GUS at 4-dpi, consistent with the induction of GFP silencing with a red ring observed (Fig. 3A,B). Notably, the GFP mRNA level was even lower in leaves co-infiltrated with 35S-PSTVd or 35S-Virp1 compared to leaves co-infiltrated with 35S-GUS at this time point (Fig. 3B). This result demonstrates that the PSTVd replication increased silencing of GFP mRNA was, at least in part, attributable to the functions of Virp1, whose mRNA was induced by PSTVd replication (Fig. 2). Low levels of siGFP were again detected in leaves co-infiltrated with 35S-GFP/35S-PSTVd but not in 35S-GFP/35S-Virp1 compared to control 35S-GFP/35S-GUS co-infiltration (Fig. 3B). These results suggest that, in addition to the degradation at the PTGS level, the enhanced silencing of GFP by replicative PSTVd or Virp1 was reinforced at the TGS level by altering the GFP mRNA expression level.
Effects of PSTVd replication and Virp1 on the expression and DNA methylation of the GFP transgene.
(A) GFP fluorescence in leaves of 16c plants co-infiltrated with 35S-GFP and either 35S-GUS control, 35S-PSTVd or 35S-Virp1. Photographs were taken under UV light at 4 dpi. (B) RNA gel blot detection of GFP mRNA, mature PSTVd RNA and siGFP accumulation from samples as described in (A). Methylene blue-stained ribosome rRNA and U6 hybridization were used as the loading control. (C) Percentage of CG, CHG, and CHH methylation of the 35S promoter of the 35S-GFP transgene by bisulfite sequencing analysis from samples as described in (A). The original data are shown in the Supplementary Figure S4. The statistical analysis was performed using the OriginPro 8 program.
As DNA methylation at the promoter sequences can cause TGS20, we investigated if DNA methylation occurred in the 35S promoter of the 35S-GFP transgene using bisulfite sequencing. DNA samples were extracted from leaves co-infiltrated with 35S-GFP and 35S-PSTVd, 35S-Virp1 or 35S-GUS. As summarized in Fig. 3C, the sequencing results revealed increases in methylation of CG, CHG and CHH in the 35S promoter region of 35S-PSTVd-co-infiltrated leaves compared to the35S-GUS-co-infiltrated leaves. In the 35S-Virp1-co-infiltrated leaves, an increased ratio of DNA methylation was also detected compared to that of 35S-GUS-co-infiltrated leaves, demonstrating that expression of Virp1 also contributed to the changes of DNA methylation. The increased ratio of DNA methylation in 35S-PSTVd-co-infiltrated leaves was clearly higher than in 35S-Virp1-co-infiltrated leaves, suggesting that, in addition to Virp1, other factor(s) may also be involved in the increases in DNA methylation during PSTVd replication in plants.
Taken together, our data demonstrate that PSTVd replication caused the reduced expression of the GFP transgene and the increased DNA methylation of its promoter, a process that is partially dependent on the functions of Virp1, which is increased upon replication of PSTVd.
PSTVd infection induced Virp1 mRNA and increased DNA methylation in host potato plants
Next, we investigated whether PSTVd infection in natural host potato induced Virp1 mRNA. Potato plants were inoculated with sap extracted from natural PSTVd-infected potato plants. Typical symptoms were observed in infected plants with a decrease in shoot and internode length, shortening of petioles and distortion of leaves (Fig. 4A). Replication of PSTVd in infected potato was detected in both inoculated and systemic leaves (Fig. 4B). The high level of viroid detected in inoculated leaves probably included the initial inoculums. TheVirp1 mRNA expression level was induced in both inoculated and, especially, systemic leaves (Fig. 4B). This result demonstrates that PSTVd infection in the host potato plants also greatly increased Virp1 mRNA expression. microRNAs are known to control the expression of genes involved in several developmental processes. To detect whether PSTVd infection affects the host miRNA pathway, the accumulation of conserved miRNAs was examined. There were no significant differences in miR159 and miR167 accumulation in potato plants before and after PSTVd infection (Fig. 4C). siPSTVd was detected in PSTVd-infected potato plants (Fig. 4C). These data demonstrate that replication of viroid RNA in potato also triggers RNA silencing and greatly induces Virp1 mRNA. PSTVd infection and increases in Virp1 transcripts do not significantly affect the miRNA pathway in potato plants.
Effects of PSTVd infection on Virp1 expression and DNA methylation in host potato plants.
(A) Disease symptoms on potato plants inoculated with PSTVd at 25-dpi and 45-dpi. (B) RNA gel blot detection of Virp1 mRNA accumulation and Dot-blot hybridization detection of PSTVd accumulation in both inoculated and systemic leaves of the host potato plants. Methylene blue-stained ribosome rRNA was used as the loading control. (C) RNA gel blot analysis of endogenous small RNAs and siPSTVd in PSTVd-infected host potato plants. U6 hybridization was used as the loading control. (D) Quantitative RT-PCR analysis of the expression levels of the selected genes in PSTVd-infected host potato plants. Error bars represent SD for three replicates. Relative transcript levels were calculated by the ∆∆C(t) method (Livak and Schmittgen, 2001) using EF1-a transcripts (potato elongation factor 1−α) as the internal standard. The value of mRNA in plants (−) was arbitrarily designated as 1. The asterisks indicate significant differences (P < 0.05, one-way ANOVA). McrBC PCR analysis of the DNA methylation levels of selected gene promoter sequences in PSTVd-infected host potato plants.
We then examined whether PSTVd-infected potato would have an effect on DNA methylation. Four sequenced genes with a high cytosine context in promoters were selected to examine expression levels and DNA methylation. Quantitative RT-PCR analysis demonstrated that the expression of the selected genes decreased in PSTVd-infected potato plants compared to the mock inoculation wild-type plants (Fig. 4D). We then investigated the methylation state of the selected gene promoter sequences using McrBC digestion-PCR. McrBC is a methylation-dependent restriction enzyme that recognizes DNA containing two or more methylated cytosine residues and cleaves the DNA at multiple sites close to one of the methylated cytosines. PCR-amplified McrBC-digested DNA resulted in a clear reduction in the amplification of four tested gene promoters in PSTVd-infected potato plants, whereas there was no significant difference in the amplification of the EF1-α gene promoter which contains a low level of cytosine residues (Fig. 4D). This indicated that the tested gene promoters were hypomethylated in PSTVd-infected plants. Taken together, consistent with the finding of PSTVd-induced Virp1-associated expediting GFP silencing in N. benthamiana plants, PSTVd infection in natural host potato plants also increased Virp1 transcript and DNA methylation in promoters of certain endogenous genes resulting in reduced transcription levels.
Discussion
In this study, we showed that PSTVd infection in N. benthamiana and its natural host potato plants triggered RNA silencing and produced a large amount of siPSTVd. However, unlike many viruses encoding suppressor proteins (VSR) to counter host anti-viral RNA silencing, we found that replication of PSTVd accelerated GFP silencing in a transient expression assay by increasing TGS via enhancing target promoter DNA methylation. The Virp1 coding gene was required for and induced by PSTVd replication and, in part, contributed to promoter methylation of the GFP transgene. Consistently, PSTVd infection in natural host potato also induced Virp1 mRNA and increased promoter DNA methylation of certain endogenous genes.
Lacking protein-coding capacity, interference with DNA methylation by PSTVd must direct interaction of its genomic RNA or derivatives with host factors. It has recently been reported that infection of cucumber with a nuclear-replicating HSVd is accompanied by dynamic changes in DNA methylation of host ribosomal (rb)RNA genes, likely associated with alterations in the levels of endogenous rb-sRNAs35. However, host factors involved in changing DNA methylation have not been identified. Consistent with previous findings that the PSTVd interaction with Virp1 was required for PSTVd infection in both Nicotiana plants and protoplasts32,33, in this study, we demonstrate that silencing of Virp1 by an RNAi construct prevented PSTVd replication in N. benthamiana plants. Virp1 is a bromodomain-containing protein with an atypical RNA binding domain and a nuclear localization signal. Bromodomain-containing proteins have the “chromatin association mark” and have been recognized as a new partnership for silencing37,38. For example, SWI/SNF is a well-characterized chromatin remodeling complex, and the Swi2/snf2 bromodomain is important for SWI/SNF-mediated displacement of acetylated histones in Yeast and Hela nucleosomes39,40. Two plant-specific subfamilies of SNF2 domain-containing proteins, CLSY1 and DRD1, are implicated in DNA methylation and nuclear RNA silencing, along with Pol IV and Pol V, respectively41,42,43. Moreover, DRD1 could contribute to the dynamic regulation of DNA methylation42, as Virp1 is also a SNF2-like subfamily protein and its carboxy-terminal RNA-binding domain has been shown to interact specifically with the right terminal domain of the viroid RNA in vivo and in vitro32,33. Our results show that PSTVd replication in both N. benthamiana and potato plants induced Virp1 and enhanced gene silencing by interfering with DNA methylation. We propose that the Virp1 protein itself may play an additional role in RNA-mediated methylation, in addition to its requirement for PSTVd infection. Alternatively, PSTVd may possess the capability to interact with other SNF2 domain-containing protein(s), such as CLSY1 and DRD1, which are associated with nuclear RNA silencing. Therefore, PSTVd replication in the nucleus may interfere with the nuclear silencing pathway, resulting in the alteration of DNA methylation. In fact, the lower effect of Virp1 than PSTVd on DNA methylation suggests that other factors are involved in or affected by PSTVd replication and might also contribute to accelerating the silencing process. It has been reported that DNA-dependent RNA polymerase II (Pol II) is implicated in PSTVd transcription28,29,30,31, and that Pol II coordinates Pol IV and Pol V in TGS and RdDM in Arabidopsis44,45,46. Thus, subunits of Pol II that are implicated in PSTVd transcription could also be possible candidates involved in the alteration of DNA methylation during PSTVd infection. Our results provide new insights into the plant-viroid interaction on viroid pathogenicity by subverting the plant cell silencing machinery.
Viroid molecules are presumed to exist in their native state in the cell as rod-like structures, characterized by a series of short double helices and internal loops resulting from intramolecular base-pairing27,47, with a structural similarity to the host’s imperfect stem-loop miRNA precursors, which look similar to the terminal parts of viroids. Although microRNAs are known to control the expression of genes involved in several developmental processes48,49,50,51, our data showed that PSTVd infection was unlikely to affect the biosynthesis of endogenous miRNAs, even though potato plants infected with PSTVd showed alteration in leaf morphology and growth, consistent with the previous report that Citrus exocortis viroid (CEVd)-infected tomato did not affect the endogenous miRNA pathway17. In fact, it has been shown that an artificial miRNA that corresponds to sequences within the PSTVd virulence modulating region (VMR) induced abnormal phenotypes in Nicotiana tabacum and N. benthamiana that closely resemble those displayed by PSTVd-infected plants52, suggesting that PSTVd-derived siRNAs could direct RNA silencing of targeted host gene(s). This could also account for viroid pathogenicity53.
The PSTVd in its native state is an effective DCL substrate but is largely inaccessible by the RISC complex26. It is likely that the DCL-mediated primary viRNAs are essential but not sufficient for antiviral defense14. We presume that the increase in expression of Virp1 upon PSTVd has a dual role in the PSTVd-host interaction. On the one hand, PSTVd replication requires Virp1. On the other hand, the RNA binding domain and nuclear localization signal of Virp1 facilitate viroid cutting into viRNAs by DCL proteins located in the nucleus. This may compensate for the inefficient RISC-associated anti-PSTVd in viroid replicating plant cells.
In summary, in this study, we demonstrate a novel activity of a subviral silencing enhancer in targeting DNA methylation associated with host Virp1, a bromodomain-containing protein. To the best of our knowledge, this is the first demonstration of involvement of a host factor in interfering with DNA methylation induced by a pathogenic non-coding viroid RNA. We cannot rule out that PSTVd replication would also cause reduction of host genome DNA methylation during infection, as recently reported for HSVd infection in cucumber for dynamic alteration of rbDNA35. Genome-wide analyses of the DNA methylome in PSTVd-infected plants would help to define the impact of PSTVd infection on DNA methylation and host gene transcription in viroid pathogenesis.
Additional Information
How to cite this article: Lv, D.-Q. et al. Replication of a pathogenic non-coding RNA increases DNA methylation in plants associated with a bromodomain-containing viroid-binding protein. Sci. Rep. 6, 35751; doi: 10.1038/srep35751 (2016).
References
Baulcombe, D. RNA silencing. Trends Biochem Sci 30, 290–293, doi: 10.1016/j.tibs.2005.04.012 (2005).
Meister, G. & Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349, doi: 10.1038/nature02873 (2004).
Matzke, M. A. & Birchler, J. A. RNAi-mediated pathways in the nucleus. Nat Rev Genet 6, 24–35, doi: 10.1038/nrg1500 (2005).
Heo, I. & Kim, V. N. Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell 139, 28–31, doi: 10.1016/j.cell.2009.09.013 (2009).
Zhang, H. & Zhu, J. K. RNA-directed DNA methylation. Curr Opin Plant Biol 14, 142–147, doi: 10.1016/j.pbi.2011.02.003 (2011).
Ding, S. W. RNA-based antiviral immunity. Nat Rev Immunol 10, 632–644, doi: 10.1038/nri2824 (2010).
Wong, J. et al. Roles of small RNAs in soybean defense againstPhytophthora sojaeinfection. The Plant Journal 79, 928–940, doi: 10.1111/tpj.12590 (2014).
Weiberg, A. & Jin, H. Small RNAs—the secret agents in the plant–pathogen interactions. Current Opinion in Plant Biology 26, 87–94, doi: 10.1016/j.pbi.2015.05.033 (2015).
Duan, C. G., Wang, C. H. & Guo, H. S. Application of RNA silencing to plant disease resistance. Silence 3, 5, doi: 10.1186/1758-907x-3-5 (2012).
Aliyari, R. et al. Mechanism of Induction and Suppression of Antiviral Immunity Directed by Virus-Derived Small RNAs in Drosophila. Cell Host & Microbe 4, 387–397, doi: 10.1016/j.chom.2008.09.001 (2008).
Baumberger, N., Tsai, C. H., Lie, M., Havecker, E. & Baulcombe, D. C. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17, 1609–1614, doi: 10.1016/j.cub.2007.08.039 (2007).
Díaz-Pendón, J. A. & Ding, S.-W. Direct and Indirect Roles of Viral Suppressors of RNA Silencing in Pathogenesis. Annual review of phytopathology 46, 303–326, doi: 10.1146/annurev.phyto.46.081407.104746 (2008).
Guo, H. S. & Ding, S. W. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J 21, 398–407, doi: 10.1093/emboj/21.3.398 (2002).
Zhao, J.-H., Hua, C.-L., Fang, Y.-Y. & Guo, H.-S. The dual edge of RNA silencing suppressors in the virus–host interactions. Current Opinion in Virology 17, 39–44, doi: 10.1016/j.coviro.2015.12.002 (2016).
Itaya, A., Folimonov, A., Matsuda, Y., Nelson, R. S. & Ding, B. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol Plant Microbe Interact 14, 1332–1334, doi: 10.1094/MPMI.2001.14.11.1332 (2001).
Wang, M. B. et al. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc Natl Acad Sci USA 101, 3275–3280 (2004).
Martín, R. et al. Characterization of small RNAs derived from Citrus exocortis viroid (CEVd) in infected tomato plants. Virology 367, 135–146, doi: 10.1016/j.virol.2007.05.011 (2007).
Du, Q. S. et al. DCL4 targets Cucumber mosaic virus satellite RNA at novel secondary structures. J Virol 81, 9142–9151, doi: 10.1128/JVI.02885-06 (2007).
Denti, M. A., Boutla, A., Tsagris, M. & Tabler, M. Short interfering RNAs specific for potato spindle tuber viroid are found in the cytoplasm but not in the nucleus. Plant J 37, 762–769 (2004).
Zahid, K. et al. Nicotiana Small RNA Sequences Support a Host Genome Origin of Cucumber Mosaic Virus Satellite RNA. PLoS Genetics 11, e1004906, doi: 10.1371/journal.pgen.1004906 (2015).
Hou, W. N., Duan, C. G., Fang, R. X., Zhou, X. Y. & Guo, H. S. Satellite RNA reduces expression of the 2b suppressor protein resulting in the attenuation of symptoms caused by Cucumber mosaic virus infection. Molecular plant pathology 12, 595–605 (2011).
Zhu, H. et al. Satellite RNA-derived small interfering RNA satsiR-12 targeting the 3′ untranslated region of Cucumber mosaic virus triggers viral RNAs for degradation. J Virol 85, 13384–13397 (2011).
Su, X., Fu, S., Qian, Y., Xu, Y. & Zhou, X. Identification of Hop stunt viroid infecting Citrus limon in China using small RNAs deep sequencing approach. Virology journal 12, 103, doi: 10.1186/s12985-015-0332-2 (2015).
Adkar-Purushothama, C. R., Perreault, J. P. & Sano, T. Analysis of small RNA production patterns among the two potato spindle tuber viroid variants in tomato plants. Genom Data 6, 65–66, doi: 10.1016/j.gdata.2015.08.008 (2015).
Gomez, G. & Pallas, V. Mature monomeric forms of Hop stunt viroid resist RNA silencing in transgenic plants. Plant J 51, 1041–1049, doi: 10.1111/j.1365-313X.2007.03203.x (2007).
Itaya, A. et al. A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. Journal of Virology 81, 2980–2994 (2007).
Flores, R., Hernandez, C., Martinez de Alba, A. E., Daros, J. A. & Di Serio, F. Viroids and viroid-host interactions. Annual review of phytopathology 43, 117–139, doi: 10.1146/annurev.phyto.43.040204.140243 (2005).
Daros, J. A., Elena, S. F. & Flores, R. Viroids: an Ariadne’s thread into the RNA labyrinth. EMBO reports 7, 593–598, doi: 10.1038/sj.embor.7400706 (2006).
Qi, Y. & Ding, B. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15, 1360–1374 (2003).
Wang, Y. et al. A Land Plant-Specific Transcription Factor Directly Enhances Transcription of a Pathogenic Noncoding RNA Template by DNA-Dependent RNA Polymerase II. The Plant Cell 28, 1094–1107, doi: 10.1105/tpc.16.00100 (2016).
Flores, R., Gas, M. E., Molina, D., Hernandez, C. & Daros, J. A. Analysis of viroid replication. Methods in molecular biology (Clifton, N.J) 451, 167–183, doi: 10.1007/978-1-59745-102-4_12 (2008).
Maniataki, E., Martinez de Alba, A. E., Sagesser, R., Tabler, M. & Tsagris, M. Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. Rna 9, 346–354 (2003).
Kalantidis, K. et al. Virp1 Is a Host Protein with a Major Role in Potato Spindle Tuber Viroid Infection in Nicotiana Plants. Journal of Virology 81, 12872–12880, doi: 10.1128/jvi.00974-07 (2007).
Pelissier, T. & Wassenegger, M. A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. Rna 6, 55–65 (2000).
Martinez, G., Castellano, M., Tortosa, M., Pallas, V. & Gomez, G. A pathogenic non-coding RNA induces changes in dynamic DNA methylation of ribosomal RNA genes in host plants. Nucleic acids research 42, 1553–1562, doi: 10.1093/nar/gkt968 (2014).
Guo, H. S., Fei, J. F., Xie, Q. & Chua, N. H. A chemical-regulated inducible RNAi system in plants. Plant J 34, 383–392 (2003).
Yang, X. J. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 26, 1076–1087, doi: 10.1002/bies.20104 (2004).
Daniel, J. A., Pray-Grant, M. G. & Grant, P. A. Effector proteins for methylated histones - An expanding family. Cell Cycle 4, 919–926, doi: 10.4161/Cc.4.7.1824 (2005).
Hassan, A. H. et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111, 369–379 (2002).
Hassan, A. H. et al. Selective recognition of acetylated histones by bromodomains in transcriptional co-activators. Biochem J 402, 125–133, doi: 10.1042/bj20060907 (2007).
Kanno, T. et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nature Genetics 37, 761–765, doi: 10.1038/ng1580 (2005).
Kanno, T. et al. A SNF2-like protein facilitates dynamic control of DNA methylation. EMBO reports 6, 649–655, doi: 10.1038/sj.embor.7400446 (2005).
Smith, L. M. et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19, 1507–1521 (2007).
Zheng, B. et al. Intergenic transcription by RNA Polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev (2009).
Gao, Z. et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465, 106–109, doi: 10.1038/nature09025 (2010).
He, X. J. et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev 23, 318–330, doi: 10.1101/gad.1765209 (2009).
Tsagris, E. M., Martinez de Alba, A. E., Gozmanova, M. & Kalantidis, K. Viroids. Cellular microbiology 10, 2168–2179, doi: 10.1111/j.1462-5822.2008.01231.x (2008).
Rhoades, M. W. et al. Prediction of plant microRNA targets. Cell 110, 513–520, doi: S0092867402008632 (2002).
Palatnik, J. F. et al. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263, doi: 10.1038/Nature01958 (2003).
Jones-Rhoades, M. W., Bartel, D. P. & Bartel, B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57, 19–53, doi: 10.1146/annurev.arplant.57.032905.105218 (2006).
Wang, J.-J. & Guo, H.-S. Cleavage ofINDOLE-3-ACETIC ACID INDUCIBLE28mRNA by MicroRNA847 Upregulates Auxin Signaling to Modulate Cell Proliferation and Lateral Organ Growth in Arabidopsis. The Plant Cell 27, 574–590, doi: 10.1105/tpc.15.00101 (2015).
Eamens, A. L., Smith, N. A., Dennis, E. S., Wassenegger, M. & Wang, M.-B. In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of Potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology 450–451, 266–277, doi: 10.1016/j.virol.2013.12.019 (2014).
Flores, R., Owens, R. A. & Taylor, J. Pathogenesis by subviral agents: viroids and hepatitis delta virus. Curr Opin Virol 17, 87–94, doi: 10.1016/j.coviro.2016.01.022 (2016).
Acknowledgements
This work was supported by the China Postdoctoral Science Foundation (20110491125), the Natural Science Foundation of China (31471753, 31501608) and Heilongjiang Province and CAS Cooperation Project (HZ201312).
Author information
Authors and Affiliations
Contributions
Y.-Y.F., D.-Q.L. and H.-S.G. conceived the study and designed the research. D.-Q.L., S.-W.L. and Y.-Y.F. performed molecular work. S.-W.L. and J.-H.Z. performed the sampling and analyzed the data. B.-J.Z. and S.-P.W. assisted construction and infection. Y.-Y.F., H.-S.G. and J.-H.Z. wrote the manuscript. All authors have read and approved the final version of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lv, DQ., Liu, SW., Zhao, JH. et al. Replication of a pathogenic non-coding RNA increases DNA methylation in plants associated with a bromodomain-containing viroid-binding protein. Sci Rep 6, 35751 (2016). https://doi.org/10.1038/srep35751
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep35751
This article is cited by
-
Phytopathogen-induced changes to plant methylomes
Plant Cell Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.