Abstract
Knockin (KI) mouse carrying a point mutation has been an invaluable tool for disease modeling and analysis. Genome editing technologies using the CRISPR/Cas system has emerged as an alternative way to create KI mice. However, if the mice carry nucleotide insertions and/or deletions (InDels) in other genes, which could have unintentionally occurred during the establishment of the KI mouse line and potentially have larger impact than a point mutation, it would confound phenotyping of the KI mice. In this study, we performed whole exome sequencing of multiple lines of F1 heterozygous Ntrk1 KI mice generated using the CRISPR/Cas system in comparison to that of a wild-type mouse used as a control. We found three InDels in four KI mice but not in a control mouse. In vitro digestion assay suggested that each InDel occurred as a de novo mutation, was carried-over from the parental mice, or was incorporated through the Cas9 nuclease mediated off-target cleavage. These results suggest that frequency of InDels found in KI mice generated by the CRISPR/Cas technology is not high, but cannot be neglected and careful assessment of these mutations is warranted.
Similar content being viewed by others
Introduction
Knockin (KI) mice harboring point mutations that are observed in genomic DNA in patients or those that alter crucial amino acids/nucleotides of proteins/genes have been widely used for investigating human diseases as well as for analyzing gene function. One of the most popular approaches for the development of these mice utilizes mouse embryonic stem (ES) cells with a homologous recombination to introduce changes in the genome1. Recently, genome editing technologies such as Zinc finger nuclease (ZFN), TALE nuclease (TALEN), and the CRISPR/Cas RNA-guided nuclease system have emerged as easy and highly efficient methods to this end2,3,4.
In the CRISPR/Cas system, upon introduction into cells, the Cas9 nuclease targets the genomic DNA by a single-guide RNA (sgRNA) consisting of a 20-nucleotide guide sequence and a scaffold5. The guide sequence pairs to the target DNA region that lies upstream of the 5’-NGG protospacer adjacent motif (PAM). Cas9 causes a double-strand break (DSB) ~3 bp upstream of the PAM in a highly specific and efficient manner, and the DSB is repaired by non-homologous end-joining, leading to nucleotide insertions and/or deletions (InDels). However, Cas9 can mediate DSBs for targets with similar sequences, which is referred to as its off-target effects6,7. To avoid off-target effects, Cas9 nickase, a modified version of Cas9 that cleaves only one strand, can be used with a pair of nearby sgRNAs8,9 to generate knock out (KO) mice10. In generating KI mice, however, the success rate is much lower than that in simple KO mice11. Though Cas9 nickase enables precise digestion, its efficiency to cleave DNA is compromised. In addition, to introduce a point mutation at a specific site, it would not be easy to design a pair of sites for sgRNAs surrounding the specific cleavage site12. Thus, in generating KI mice, wild-type Cas9 having high efficiency of DNA cleavage, is usually used. Therefore, a possibility that unintended mutations occur cannot be neglected.
In studying behavioral phenotypes of KI mice carrying a single missense mutation generated using the CRISPR/Cas system, it is assumed that the phenotype of KI mice should be milder than knockout mice in general. Thus, phenotyping the KI mice is based on assumption that there are no other InDels that perturb the gene function. This would especially be important when looking for subtle behavioral changes in mouse models of mental disorders carrying a single missense mutation. To avoid the possible effects of other InDels in KI mice generated using CRISPR/Cas, researchers can adopt several strategies; 1) checking the off-target sites predicted using software, 2) back-crossing with wild-type mice for several generations, 3) use of at least two independent KI mouse lines or sgRNAs, and 4) extensive exome/whole genome sequencing to identify the best line that does not contain other InDels. Approach 1 might overlook mutations introduced outside the predicted sites, whereas approaches 2 and 3 are time consuming. Whole genome sequencing can reveal unintended mutations in both coding and non-coding regions but is expensive. Whole exome sequencing can reveal mutations in coding regions at a reasonable cost. However, it is not known whether unintended InDels are indeed found in KI mice generated using the CRISPR/Cas technology.
Few studies have addressed unintended mutations in mutant mice generated by CRISPR/Cas. Iyer et al. carried out whole-genome sequencing (WGS) in the F1 heterozygous Androgen receptor (Ar) gene KO mice produced using CRISPR/Cas13 and found several InDels. They interpreted that the InDels mostly occurred spontaneously and not due to Cas9 nuclease activity mainly because their sequences were unrelated to predicted off-target sites. Although they validated some InDels identified, they did not investigate the cause of those InDels.
In this study, we performed whole exome analysis of F1 KI mice heterozygous for Ntrk1, a neurotrophic tyrosine kinase receptor gene, generated using the CRISPR/Cas to identify the unintended InDels and examined whether these mutations can be predicted in silico. We observed 0.75 InDels per mouse (three mutations per four mice), which cannot be predicted in silico. We analyzed the cause of the 3 InDels by in vitro cleavage assay as well as genotyping the F0 mice. As a result, we suggested 3 different causes for the incorporation of the unintended InDels during the generation of the KI mice. These results also suggest that exome sequencing would be useful to exclude a confounding effect of unintended InDels in the study of KI mice generated using the CRISPR/Cas system.
Results and Discussion
Generation of the KI mice
Using the CRISPR/Cas system, we generated KI mice harboring a point mutation in the mouse Ntrk1 gene to substitute the p.495 Glutamate with Lysine, which corresponded to change from c.1483 Guanine (G) to Adenine (A), as described in Fig. 1a. This mutation corresponds to a rare damaging missense mutation E492K (rs144901788) reported in humans. First, we constructed an expression vector for sgRNA and Cas9 and verified the sequence. To monitor nuclease activity, the plasmid DNA was transfected into the Neuro2A cell line. The target region was PCR amplified (482 bp) and subjected to analysis using Surveyor assay, which showed an efficient cleavage pattern, and nucleotide deletion was confirmed by Sanger sequencing after TA cloning of the amplified fragments; nucleotide deletion was observed in 3 out of the 10 clones examined (Fig. 1b).
Generation of the Ntrk1 knockin mice.
(a) Schematic of the target site at the Ntrk1 locus. In the double-stranded DNA, the PAM sequence is capitalized, and the sgRNA target is underlined. In the donor DNA, the single-strand oligodeoxynucleotide, the replaced nucleotides are capitalized (for KI, green; for creating the underlined TfiI site, red). (b) Sequence chromatograms for the target site of wild-type mice (top), Neuro2A cell line transfected with the CRISPR/Cas expression vector (middle), and heterozygous (F1) KI offspring (bottom).  , nucleotide deletion (3 bp);
, nucleotide deletion (3 bp);  ,
,  , nucleotide changes.
, nucleotide changes.
Through microinjection of the sgRNA, Cas9 mRNA, and donor DNA into 300 zygotes, in which 233 were transferred to uteri, 92 founder (F0) mice were born. DNA was extracted using proteinase K from the tail clips of the surviving 81 mice, followed by Sanger sequencing for the target region. Among the 162 alleles in total, mutations were observed in 156 alleles (96%), of which 6 alleles (4%) had the intended point mutation (KI, Fig. 1b). Mosaicism was also observed in 2 out of the 6 KI alleles, as reported previously14.
Whole exome analysis
Four F0 animals (No. 268, 283, 306, 316) carrying the intended point mutation (a KI allele) were mated with C57BL/6J (B6J) mice, and heterozygous KI offspring (F1) derived from each founder (4 males; 268-1, 283-1, 306-1, 316-2) were used for exome sequencing analysis. As a control, a B6J male mouse from the same breeding colony was also sequenced. Whole exome sequencing was performed with an enough depth (100–120x) for the above-mentioned 5 mice. Bioinformatics analysis followed by the filtering process (see Materials and Methods section) showed high-quality variants: 3 InDels (2 insertions and 1 deletion; Fig. 2a) and 21 single nucleotide variations (SNVs) among the 5 samples, in which all the InDels and 19 SNVs were confirmed by Sanger sequencing. Two of the 19 SNVs were shared among 2 KI samples (See Table 1 and Supplementary Table S1). In summary, SNVs were rare in the whole exomes and their frequencies were comparable to that in the wild-type mice, although only one control B6J mouse was examined.
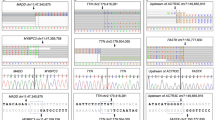
Analysis of the 3 InDels identified.
(a) Sequence information of the 3 InDel sites. Yellow highlighted nucleotides, PAM sequence for Ntrk1 target site; gray highlighted nucleotides, putative PAM sequence for Aspa insertion site. Boldface nucleotides, matched bases between the guide sequence and the Aspa sequence. (b) In vitro cleavage assay for the 3 InDel sites. Cleavage templates: PCR products amplified from wild-type B6J mouse genomic DNA including the sites for Ntrk1 target, InDels (Zfp365, Alox12, Aspa) and Ntrk2 genomic region corresponding to the Ntrk1 target.
We detected 3 InDels in 4 KI mice, of which two were predicted to lose protein function by a frameshift resulting in a premature stop codon. On an average, 0.75 InDels including 0.5 loss of function InDels were found in a KI mouse. Considering the size of the genome examined, our findings are consistent with those of a previous study involving whole genome sequencing in Ar KO mice, where approximately 24 InDels in the whole genome of a KO mouse were observed13. We aligned the sequences around the InDels to the guide RNA recognition sequences, but they did not correspond to the target sequence (Fig. 2a) nor matched with the potential off-target sites predicted using the CRISPR Design web server (http://crispr.mit.edu). As for SNVs, on average, 4 SNVs were found in a KI mouse exome, which is comparable to the 3 found in a B6J mouse. Although numerous reports have addressed off-target cleavage, some studies indicate that sequences not recognized as off-target sites could be changed, and SNVs have been found to increase during genome editing in cellular studies15,16,17, which might involve a cryptic mechanism18,19. However, the exact number of SNVs depends on the methodology used for SNV calling in next-generation sequencing technology20,21.
In a recent study on germline mutation rates in wild-type laboratory mice22, ~90–100 de novo SNVs were found in the whole-genome of a B6J mouse, which accumulated in specific lines. Here ~3 SNVs were found in the exome by simple arithmetic, which almost corresponds to the number observed in our B6J sample (Table 1). Therefore, the 3 SNVs in the B6J mouse would have naturally occurred and resulted in the accumulation of mutations in the specific B6J line. Further, the number is almost the same as that observed in CRISPR/Cas-mediated KI mice. Therefore, we cannot conclude whether the number of SNVs increased during the process of KI mouse development. For InDels, it was reported that 3–4 de novo InDels were observed in the whole genome of a B6J mouse22. This means that there would be less than 0.1 de novo InDels in the exome of B6J. Thus, an average of 0.8 exonic InDels observed in the KI mice is much higher than expected and it would be possible that this was a result of incorporation during the generation of the KI mice.
For further reference, we scrutinized the previous WGS results for F1 heterozygous KO mice generated using the CRISPR/Cas13. They performed WGS for 5 F1 mice (derived from 2 F0 mice) and 2 control mice (a B6J and a CBA mouse). They validated 22 out of the 24 InDels which was randomly selected from the 120 InDels specifically identified in the 5 F1 mice. We annotated the 120 InDels with ANNOVAR Documentation web server (http://annovar.openbioinformatics.org/en/latest/) and found an exonic 2 base pairs deletion causing frameshift in Pdzd2 gene, which was included in the 22 InDels validated. Because the mutation was shared among the 3 F1 mice derived from the identical F0 mouse, the InDel was supposed to be incorporated during the process of the generation of the F0 mouse.
To exclude a possibility that the 3 InDels identified were incorporated during the generation of the F1 mice, we performed Sanger sequencing for the 3 regions with the tail DNA samples of all 81 F0 mice. As a result, we independently identified the insertion within Zfp365 gene in a F0 mouse (No. 298). This mutation was the same as the insertion originally observed in the F1 316-2. We could not find any mutations for the other two genes Alox12 and Aspa except for F0 283, a parental strain for the F1 283-1 carrying these mutations. Intriguingly, the insertion within Zfp365 gene was not detected in the F0 316, a parent mouse of the F1 mouse, 316-2, carrying this insertion. In other words, the mutation not detected within somatic cells (tail clips) of a F0 mouse was identified in their F1 progeny. This suggests mosaicism, which is sometimes observed in mutant mice generated using the CRISPR/Cas system23. Collectively, these results clearly indicate that the 3 InDels were a result of incorporation during the F0 mice production.
As a mechanism for incorporation of the 3 InDels during the process of F0 mice generation, there are 3 possibilities; off-target cleavage by the Cas9 nuclease, de novo mutation, and carried-over from the parental B6J mice used for collecting the fertilized eggs. To directly assess a possibility that the 3 InDels were induced by the Cas9 nuclease activity, we performed in vitro cleavage assay for the 3 regions (Fig. 2b). We prepared 5 cleavage templates (~2 kb) that contain the 3 InDel regions, Ntrk1 target region (positive control), and Ntrk2 genomic region corresponding to the Ntrk1 target region (negative control), which were designed to produce ~1.2 and ~0.8 kb fragments when cleaved at the intended sites. After the incubation of the cleavage templates and the sgRNA with Cas9 nuclease for 2 hours, Ntrk1 templates were efficiently cleaved to ~1.2 and ~0.8 kb fragments, while templates for other regions were not. However, when the incubation time was extended to 8 hours, templates for Aspa gene were more significantly and differentially cleaved into fragments containing ~1.2 and ~0.8 kb. Because the Cas9 itself had nuclease activity, smearing was also observed for other templates (Zfp365, Alox12, and Ntrk2) at 8 hours’ incubation. The observed pattern of digestion for the Aspa templates was marked. This suggests a possible digestion by the Cas9 nuclease though the digestion pattern was much less clear than that of Ntrk1 target (Fig. 2b). For the other 2 sites (Zfp365 and Alox2), there was no evidence for cleavage by the Cas9 nuclease even after 8 hours.
Comparison between the Ntrk1 target sequence and the one around the insertion site in Aspa gene, considering the nearest 3 bases (5′-AGG) as a putative PAM sequence (Fig. 2a), shows 14 bases mismatch from the on-target Ntrk1 sequence, which was more than the number of accepted mismatch previously reported for off-target cleavage6,7. There is a case that a sequence with 13 bases mismatch was cleaved by a Cas9 nuclease due to the presence of a bulge structure in the sequence24. Under this condition, it seems unlikely that the Aspa sequence can be cleaved as efficiently as the Ntrk1 target sequence. Therefore, the result would be reasonable that the cleavage was not seen at 2 hours but was observed at 8 hours, contrary to the result of on-target Ntrk1 sequence.
With regard to the insertion in Zfp365 gene, the identical 2 base insertions were observed in independent 2 F0 mice. It is unlikely to observe two independent de novo mutations at the same site. Thus, it is supposed to be a carried-over from the parental B6J mice. However, the mutation should be a sparsely distributed one in the B6J breeding colony since it was not seen in the control B6J mouse.
The deletion in the Alox12 gene would be considered as a de novo mutation.
Thus we addressed the causes of the 3 InDels as follows; 2 bases insertion for Zfp365 gene was probably a carried-over in the B6J background, 17 bases deletion for Alox12 gene was de novo mutation, 3 bases insertion for Aspa gene was perhaps through the Cas9 nuclease activity.
In summary, we assessed nucleotide changes in the whole exome that were incorporated during the generation of KI mice using the CRISPR/Cas system. After bioinformatic filtering, followed by Sanger sequencing, we confirmed specific variants for the KI mice. We were able to elucidate the cause of the variations, but our result was derived from using only one sgRNA, which does not allow the generalization of our conclusion. Despite these limitations, our study provides evidence indicating that InDels, as a result of off-target cleavage by the Cas9 nuclease, can occur in KI mice produced using the CRISPR/Cas system. Further attempts to validate genetic integrity will be required when using KI mice created using CRISPR/Cas technology.
Materials and Methods
Preparation of sgRNA, Cas9 mRNA, and donor DNA
The sequence of the sgRNA was determined using CRISPR Design web server by submitting the target sequence of the mouse Ntrk1 gene. A pair of oligonucleotides for the targeting site was ligated into the pX330 (Addgene) vector5: For, 5′-caccgACTGTGGGTTCTCCATGATG-3′; Rev, 5′-aaacCATCATGGAGAACCCACAGTc-3′. For in vitro synthesis of sgRNA and Cas9 mRNA, the MEGAshortscript Kit and mMESSAGE mMACHINE T7 Ultra Kit (Life Technologies) were used, respectively4. Donor DNA, single strand oligodeoxynucleotides (120 bp), was manually designed and PAGE-purified products were purchased (Sigma): 5′-GGGTGGCAGTTCTCTTTCCCCTACTGAGGGCAAAGGCTCCGGACTCCAGGGCCACATCATGAAGAATCCACAGTACTTCAGTGATACCTGTGAGGAACTGTTATAGTAGGCGAGTGTGAG-3′.
Production of KI mice
A mixture of sgRNA, Cas9 mRNA, and the donor oligonucleotides (25 ng/μl each) was microinjected into the cytoplasm of fertilized eggs obtained from mating between B6J males and superovulated females (CLEA, Japan). The injected eggs were then transferred into the uterus of pseudopregnant ICR females.
Genotyping of the founder mice was carried out by direct sequencing of the PCR amplified fragments from the tail DNAs using the BigDye Terminator V3.1 and ABI 3730xl sequencer (Life Technologies) with the following primers: For, 5′-CTGTCAGGAGCAGGGGAGTT-3′; Rev, 5′-AGATAGGAGACAGGCTAGACTTTGA-3′. The selected founder mice were then mated with B6J mice, and heterozygous KI mice (F1) were identified in the offspring, which were then subjected to exome sequencing.
All animal experiment protocols were approved by the Wako Animal Experiment Committee, RIKEN, and all experiments were performed in accordance with the approved guidelines and regulations.
All other experimental procedures were approved by the RIKEN Wako Safety Center and were carried out in accordance with the approved guidelines.
DNA preparation and whole exome sequencing
Genomic DNAs were purified from mouse tail samples using GenElute Mammalian Genomic DNA Miniprep kit (Sigma). Exome sequencing and bioinformatics analyses were performed at RIKEN GENESIS CO., LTD.
Target capture for the exome was performed on each sample using SureSelectXT Mouse All Exon kit (Agilent Technologies). DNA was subjected to SureSelectXT Target Enrichment System for paired-end DNA library preparation. The length of the library, including adaptors, was 294–310 bp. Whole exome sequencing was performed on HiSeq2500. Sequencing data were generated with 101 bp paired-end reads.
Mapping, variant calling, and validation
Sequencing reads were aligned to the mouse reference (mm10) using BWA 0.7.10. After excluding chimeric reads, the duplicated reads were eliminated using Picard. GATK25 ‘IndelRealigner’ and ‘Table Recalibration’ were used for local realignment and for recalibrating the quality scores, respectively. For SNV/InDel calling in multi-sample analysis, GATK ‘UnifiedGenotyper’ was used for comparison with the reference genome. For SNV calling in matched-pair analysis, SomaticSniper was used to compare the difference between each KI mouse and the B6J mouse. Annotation for all variants was made using dbSNP138, CCDS (NCBI, release 20131209), and RefSeq (UCSC Genome Browser, dumped 20131124).
The raw calling result was annotated and further refined to the 1st_selection list (1,251 variants), which includes non-synonymous (1,102 missense, 37 nonsense, 3 insertion, 4 deletion, 24 frameshift) and splice site (51 splice donor or 30 splice acceptor site) mutations. These variants were further filtered using the following conditions. (1) All variants in a gene were excluded from further analysis in the case that 3 or more different variants were observed in the same gene. (2) “Known” variants were excluded to focus on novel mutations. (3) Variants with less than 10 reads in all samples were excluded as the reliability was insufficient. (4) Variants shared among B6J mice were excluded as they were supposed to be derived from a B6J mating pair. (5) Homozygous variants were excluded as the mice were from the F1 progeny. (6) Variants with ratio less than 0.5 between the numbers of mutated reads and those of non-mutated ones were also excluded.
Sanger sequencing was performed to confirm the selected variants. PCR primers were designed using GENETYX software (GENETYX Corporation).
In vitro cleavage assay
The assay was performed using Guide-it Complete sgRNA Screening System (Takara Bio). Briefly, Cas9 proteins (50 ng/μl) and in vitro transcribed Ntrk1 sgRNA (10 ng/μl) were incubated with each cleavage template (35 ng/μl, PCR amplified fragments for 5 target regions) in a Cas9 nuclease reaction buffer at 37 °C for 2 or 8 hours. Reactions were stopped by incubating at 70 °C for 10 minutes and analyzed with gel electrophoresis (1% agarose). For amplification of the cleavage templates, following primer pairs were used for each site. Ntrk1 (1949 bp), For, 5′-AGTCACCTGGCGGACAAGCAAGGA-3′, Rev, 5′-TCTCCTTCTCGCCAGTGGGTGAGTT-3′; Zfp365 (1955 bp), For, 5′-TACTAGAACCTCCTGCTCTTGAC-3′, Rev, 5′-GCTTTAACCCAAGAAGGCTGA-3′; Alox12 (1990 bp), For, 5′-GCGCCATTGAGGAACTGGTAGCCAA-3′, Rev, 5′-GGGACAAGTGCAGAGGCCGTGTTTC-3′; Aspa (1907 bp), For, 5′-CACCCGTTTCTTCTCCTTTCC-3′, Rev, 5′-ATTTGCGTGTAAGAGACAGAAGAG-3′; Ntrk2 (1951 bp), For, 5′-CATATGTTCCTGGTGATTGACTGAC-3′, Rev, 5′-TCTCATGCCATATAACAAACTCAGG-3′.
Additional Information
How to cite this article: Nakajima, K. et al. Exome sequencing in the knockin mice generated using the CRISPR/Cas system. Sci. Rep. 6, 34703; doi: 10.1038/srep34703 (2016).
References
Menke, D. B. Engineering subtle targeted mutations into the mouse genome. Genesis 51, 605–618 (2013).
Cui, X. et al. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat. Biotechnol. 29, 64–67 (2011).
Wefers, B. et al. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc. Natl. Acad. Sci. 110, 3782–3787 (2013).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/cas-mediated genome engineering. Cell 153, 910–918 (2013).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Cong, L. et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science (80-.). 339, 819–823 (2013).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Shen, B. et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399–402 (2014).
Lee, A. Y. & Lloyd, K. C. K. Conditional targeting of Ispd using paired Cas9 nickase and a single DNA template in mice. FEBS Open Bio 4, 637–642 (2014).
Singh, P., Schimenti, J. C. & Bolcun-Filas, E. A Mouse Geneticist’s Practical Guide to CRISPR Applications. Genetics 199, 1–15 (2014).
Iyer, V. et al. Off-target mutations are rare in Cas9-modified mice. Nat. Methods 12, 479 (2015).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/cas-mediated genome engineering. Cell 154, 1–10 (2013).
Smith, C. et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15, 12–13 (2014).
Suzuki, K. et al. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell 15, 31–36 (2014).
Veres, A. et al. Low incidence of Off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15, 27–30 (2014).
Gabriel, R. et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 29, 816–823 (2011).
Pattanayak, V., Ramirez, C. L., Joung, J. K. & Liu, D. R. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 8, 765–770 (2011).
Goldstein, D. B. et al. Sequencing studies in human genetics: design and interpretation. Nat. Rev. Genet. 14, 460–470 (2013).
Young, M. A. et al. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell 10, 570–582 (2012).
Uchimura, A. et al. Germline mutation rates and the long-term phenotypic effects of mutation accumulation in wild-type laboratory mice and mutator mice. Genome Res. 25, 1125–1134 (2015).
Mianné, J. et al. Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediated homology directed repair. Genome Med. 8, 16 (2016).
Wang, X. et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 33, 175–178 (2015).
McKenna, A. et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Acknowledgements
We thank Dr. Tomomi Aida (Tokyo Medical and Dental University) for technical advice. We are grateful to Research Resources Center at RIKEN Brain Science Institute for technical supports. We also thank Ms. Naoko Kume and all other members of our laboratory for their support, assistance, and the discussions. T.K.’s work has been funded by Grants-in-Aid from the Ministry of Health, Labour and Welfare, and the Ministry of Education, Culture, Sports, Science and Technology of Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. T.K. received honoraria for lectures, manuscripts, and/or consultancy, from Kyowa Hakko Kirin Co., Ltd., Eli Lilly Japan K.K. Otsuka Pharmaceutical Co., Ltd., GlaxoSmithKline K.K., Taisho Toyama Pharmaceutical Co., Ltd., Dainippon Sumitomo Pharma Co., Ltd., Meiji Seika Pharma Co., Ltd., Pfizer Japan Inc., Mochida Pharmaceutical Co., Ltd., Shionogi & Co., Ltd., Janssen Pharmaceutical K.K. Yoshitomiyakuhin, Agilent Technologies, Astellas Pharma Inc., and Wako Pure Chemical Industries, Ltd. within the last 3 years. T.K. also received a research grant from Takeda Pharmaceutical Co., Ltd outside of this work.
Author information
Authors and Affiliations
Contributions
K.N. and T.K. conceived the project and wrote the paper. K.N. and A.K. performed experiments. M.N. and T.T. assisted the experiments. J.K. assisted the conception of the project. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Nakajima, K., Kazuno, Aa., Kelsoe, J. et al. Exome sequencing in the knockin mice generated using the CRISPR/Cas system. Sci Rep 6, 34703 (2016). https://doi.org/10.1038/srep34703
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep34703
This article is cited by
-
Bioinformatic and literature assessment of toxicity and allergenicity of a CRISPR-Cas9 engineered gene drive to control Anopheles gambiae the mosquito vector of human malaria
Malaria Journal (2023)
-
Blunted Amphetamine-induced Reinforcing Behaviors and Transporter Downregulation in Knock-in Mice Carrying Alanine Mutations at Threonine-258 and Serine-259 of Norepinephrine Transporter
Journal of Molecular Neuroscience (2022)
-
Ntrk1 mutation co-segregating with bipolar disorder and inherited kidney disease in a multiplex family causes defects in neuronal growth and depression-like behavior in mice
Translational Psychiatry (2020)
-
Comparative analysis of single-stranded DNA donors to generate conditional null mouse alleles
BMC Biology (2018)
-
Response to “Unexpected mutations after CRISPR–Cas9 editing in vivo”
Nature Methods (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.