Abstract
Alkaloid accumulation in plants is activated in response to stress, is limited in distribution and specific alkaloid repertoires are variable across taxa. Rauvolfioideae (Apocynaceae, Gentianales) represents a major center of structural expansion in the monoterpenoid indole alkaloids (MIAs) yielding thousands of unique molecules including highly valuable chemotherapeutics. The paucity of genome-level data for Apocynaceae precludes a deeper understanding of MIA pathway evolution hindering the elucidation of remaining pathway enzymes and the improvement of MIA availability in planta or in vitro. We sequenced the nuclear genome of Rhazya stricta (Apocynaceae, Rauvolfioideae) and present this high quality assembly in comparison with that of coffee (Rubiaceae, Coffea canephora, Gentianales) and others to investigate the evolution of genome-scale features. The annotated Rhazya genome was used to develop the community resource, RhaCyc, a metabolic pathway database. Gene family trees were constructed to identify homologs of MIA pathway genes and to examine their evolutionary history. We found that, unlike Coffea, the Rhazya lineage has experienced many structural rearrangements. Gene tree analyses suggest recent, lineage-specific expansion and diversification among homologs encoding MIA pathway genes in Gentianales and provide candidate sequences with the potential to close gaps in characterized pathways and support prospecting for new MIA production avenues.
Similar content being viewed by others
Introduction
The power of genomics to illuminate evolutionary hypotheses lies in the availability of sequences from both early and late branching lineages for comparative analyses. Alkaloid accumulation in plant tissues is triggered in response to the range of stress encountered by plants; for humanity the result is a pharmacopeia ranging from caffeine to heroin to some of the most important chemotherapeutic and hypertensive treatment drugs available today1. Given that plant metabolic networks have followed a descent with modification pattern2 and the unique and variable character of monoterpenoid indole alkaloid (MIA) production throughout the angiosperm order Gentianales, genome-scale comparative analyses within the order relative to more distant lineages could provide insight on the genetic factors involved in the evolution of this highly valuable aspect of plant secondary metabolism.
Alkaloid production in plants is limited in distribution and the specific alkaloid repertoire is variable across taxa. With the exception of the MIA quinolone derivatives, i.e. camptothecin, Gentianales are the exclusive plant producers of highly valuable MIA compounds, including the bisindole topoisomerase inhibitors vincristine and vinblastine. Regardless of their final form, more than 2500 identified MIAs begin with the condensation of tryptamine and secologanin to form strictosidine, the first committed molecule in the MIA pathway. The classification of MIAs into three broad groups was based on the arrangement of the fragmented cyclic monoterpene (secologanin), Iboga (catharanthine), Corynanthe-strychnos (ajmalicine) and Aspidosperma (stemmadenine). While all families of the order contain representatives that can accumulate MIA species with the Corynathe (type I) skeleton, Rauvolfioideae, a subfamily of Apocynaceae, represents a major center of structural expansion encompassing the diversity of skeletal arrangements and an astonishing number of downstream modifications yielding thousands of unique molecules3.
Rhazya stricta Decne. (2n = 22; Apocynaceae, Rauvolfioideae) is an evergreen shrub abundant across Western and South Asia and is well adapted to harsh conditions4. Like other Rauvolfioideae, Rhazya accumulates MIAs. In fact, strictosidine was first isolated from R. stricta in 19685. However recent study has focused mainly on Catharanthus roseus (L.) G. Don and, to a lesser extent, Rauvolfia serpentina (L.) Benth. ex Kurz and a handful of others in the family. There is an extensive literature describing chemical synthetic approaches to generate the kind of molecules produced by these species as, at least in the systems that have been studied, in vivo production is far below human demand6. Historical technological challenges to high quality assembly of complete nuclear genomes have resulted in fragmented drafts of limited use due to the exclusion of repetitive sequences and the lack of information on genome structure. Furthermore, the paucity of genome level data for Apocynaceae precludes a deeper understanding of MIA pathway evolution hindering the elucidation of remaining pathway enzymes and the potential to improve MIA availability in planta or in vitro. To address this, we sequenced the nuclear genome of Rhazya stricta and present this high quality assembly in comparison with that of coffee (Rubiaceae, Coffea canephora Pierre ex A. Froehner, Gentianales) and others to investigate the evolution of genome-scale features. Using genomic and transcriptomic sequences from Rauvolfioideae along with species in the order Gentianales and beyond, we identified homologs of characterized MIA pathway genes revealing the lineage specific nature of their evolution.
Results and Discussion
We sequenced three Illumina libraries yielding nearly 400 million reads to generate approximately 112X coverage of the R. stricta ~274 Mb genome. To achieve the highest quality draft with minimal fragmentation we added PacBio sequencing, (>550K reads; 10X coverage) and isolated nuclei for optical mapping (Supplementary Table S1). The final assembly contains 980 scaffolds with a maximum scaffold length of 16.4 Mb and N50 >5.5 Mb. Scaffolds of greater than 200 Kb (113) comprise 93% of the genome sequence (Supplementary Table S2, Supplementary Table S3). Comparison to assembly statistics reported for three recently published nuclear genomes demonstrates the high quality of the Rhazya draft (Supplementary Table S4). A comprehensive listing of statistics for 34 non-model plant nuclear genomes was recently reported by Unamaba et al.7.
The transcriptome-guided annotation of the R. stricta genome identified 21,164 protein-coding genes (Supplementary Table S5), 60% of which were assigned UniProt identifiers. Comparison to a set of core eukaryotic sequences (CEGMA)8 demonstrated that the R. stricta assembly captured 98% of the expected genes. The genome contains 16% transposable elements, of which 88% are long terminal repeat (LTR) retrotransposons comprising predominantly Copia and Gypsy superfamily members. Our list of repeated elements is not exhaustive and the actual value for repeat content is likely higher (Supplementary Table S6). All functionally annotated transcripts, as described in the methods section, were assigned gene names, putative functions, gene ontology (GO) terms and enzyme codes by homology search (BLAST) against the UniProt database.
A metabolic pathway database, RhaCyc, was created for Rhazya using the PathoLogic component of Pathway Tools (http://bioinformatics.ai.sri.com/ptools/). This program inferred metabolic pathways by comparing the annotated Rhazya genes to the MetaCyc 2.0 reference pathway database (http://www.metacyc.org). The RhaCyc database contains information on 3380 enzymes, 2828 compounds and 458 pathways and is available as a community resource (http://rhacyc.icmb.utexas.edu:1555/). Links to UniProt identifiers and GO terms are embedded with each annotated sequence in RhaCyc.
The high quality draft of the Coffea canephora nuclear genome9 was used to examine structural changes between the two Gentianales families and relative to the ancestral post-γ eudicot arrangement as represented by Vitis10 (Fig. 1; Supplementary Fig. S1). As in Coffea, there was no evidence to suggest a subsequent whole-genome polyploidization in the Rhazya lineage. Comparison of adjacencies within each of the Rhazya scaffolds derived from different ancestral chromosomes and those within the Coffea chromosomes allowed us to identify most of the major gene order changes in the extant genomes. While the remaining Rhazya scaffolds each tend to reflect a part of only a single ancestral chromosome, they may contain some smaller rearrangements that went undetected. With the available data we detected limited rearrangement between the core eudicot ancestor and the Gentianales ancestor and between the latter and Coffea. In the Rhazya lineage, however, many more structural rearrangements were inferred.
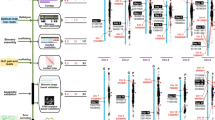
Divergent structural evolution in Rhazya stricta.
Genome level rearrangements leading from the ancestral core eudicot, represented by Vitis (upper right hand corner) to Rhazya and Coffea. Each linear arrangement represents successive steps from the eudicot ancestor toward the Gentianales ancestor. An arrow leads from the last of these stages to the relevant portion of the Gentianales ancestor. From this ancestor, two arrows indicate the divergence of Coffea and Rhazya and successive rearrangements within each lineage are again indicated by linear arrangements of colored blocks. Inferred rearrangements between lineages are given at the arrows. Extant genomes carry the species name. The circular diagrams at the lower left and right represent the current orthologies between Rhazya superscaffolds 6 and 15 and Coffea and Vitis, respectively. Syntenic blocks are colored according to the 21 ancestral core eudicot chromosomes33 and do not represent biologically significant units.
To explore the evolution of MIA pathway genes we assembled a translated subject database containing transcriptomic and genomic data for sequences from Rhazya and five additional species: two Rauvolfioideae (C. roseus, R. serpentina), C. canephora (Rubiaceae), a non-Gentianales asterid (Solanum lycopersicum L.) and an alkaloid producing rosid (Theobroma cacao L. cv Matina). For homolog identification we queried the database using previously characterized MIA protein sequences from C. roseus and R. serpentina, which produce alkaloids currently employed in cancer chemotherapy and as antiarrhythmic drugs, respectively. Putative homologs for each protein were aligned and used to generate phylogenies (Supplementary Fig. S2) for 21 genes (Supplementary Table S7). Of these, eight trees were reduced and restricted to the relevant clades as described in methods and reconciled with the species tree, depicting the history of duplication and loss for each (Figs 2 and 3, Supplementary Fig. S3, Supplementary Table S8).
Expansion, loss and duplication of selected monoterpenoid indole alkaloid (MIA) sequences across Gentianales.
The cladogram (top, center) depicts relationships among six eudicots based on the Angiosperm Phylogeny Group (http://www.mobot.org/MOBOT/research/APweb/). Histograms below each species indicate the number of sequences returned in BLAST searches to identify homologs for each of the eight gene families. The maximum likelihood trees along the left and right show subclades of reconciled gene phylogenies that include known homologs in either Catharanthus (cra.Genbank) or Rauvolfia (rsa.Genbank) for seven of the eight MIA genes (THAS is shown in Fig. 3). Complete reconciled phylogenies for each of the eight sequences are shown in Supplementary Fig. S3. cra, Catharanthus roseus; cof, Coffea canephora; rha, Rhazya stricta; tom, Solanum lycopersicum; rsa, Rauvolfia serpentina. Blue font indicates GenBank sequences and red font indicates Rhazya stricta sequences. Red square = gene duplication; green circle = speciation event; gray circles = gene loss. Values above branches indicate bootstrap support for the clade, numbers below nodes give bootstrap support for the event (duplication or speciation) at the node. Bootstrap values less than 50 are not shown; inferred loss events are indicated with an asterisk and do not have associated bootstrap values.
Duplication and loss of Tetrahydroalstonine synthase (THAS) across six angiosperms based on gene tree/species tree reconciliation.
Internal and terminal branches in black reflect the amount of change along branch; dashed gray branch extensions are present for branches whose lengths were too short to enable uniform representation with branch support and length information. cra, Catharanthus roseus; cof, Coffea canephora; rha, Rhazya stricta; rsa, Rauvolfia serpentina; tom, Solanum lycopersicum; the, Theobroma cacao. Species indicated in blue font were downloaded from GenBank and those in red are from Rhazya stricta. Red square = gene duplication; green circle = speciation event; wide gray dashed line ending with a circle = gene loss. Values above branches indicate bootstrap support for the clade, numbers below nodes give bootstrap support for the event (duplication or speciation) at the node. Bootstrap values less than 50 are not shown; inferred loss events are indicated with an asterisk and do not have associated bootstrap values. Scale bar represents number of amino acid substitutions per site. The maximum likelihood (ML) tree shown is the tree with the best ML score.
Generally, the families that include the eight characterized MIA genes underwent expansion as is seen in other gene families whose members participate in secondary metabolism2 (Supplementary Table S8). Where genome level data and therefore information about genetic loci, were available gene family expansion appears to have been facilitated by localized and/or tandem duplications. The two Gentianales taxa, Coffea and Rhazya, each contain proximally arrayed homologs. Within each species some homologs are grouped in distinct clades while others are dispersed across the phylogeny suggesting different evolutionary trajectories for these sequences. One example can be seen in Coffea. The full tree generated from sequences extracted using C. roseus Strictosidine β-glucosidase (SGD) as a query contains 157 β-glucosidase superfamily members (Supplementary Fig. S2), 17 of which are proximally arrayed on Coffea chromosome two. Of those, eight chromosome two sequences were retained in the reconciled subtree (Supplementary Fig. S3). The duplications giving rise to five glucosidases (Cc02_g27160, g27180, g27190, g27230, g27290; highlighted in brown) likely occurred prior to the divergence of Gentianales while more recent duplications gave rise to three homologs that form a clade containing Coffea sequences exclusively (Cc02_g30420, g30490, g30540; highlighted in beige).
For SGD, as for all of the experimentally confirmed MIA sequences analyzed, the C. roseus and/or R. serpentina sequences downloaded from Genbank were in derived positions in the phylograms. The clades that contained the known (query) sequences were restricted to the Rauvolfioideae, however some clades also included coffee sequences following the pattern of diversification within the order and reflecting the evolution of MIA diversity and complexity.
Our analyses placed two Rhazya sequences in the clade adjacent to the GenBank SGD sequences, along with a single sequence extracted from the Rauvolfia transcriptome (Fig. 2, Supplementary Fig. S2 and S3). However, SGD was not identified and annotated in the Rhazya genome. Looking more closely at these homologs, we aligned the Rhazya and Rauvolfia sequences within and between the clades. The Rauvolfia sequence grouping with the two Rhazya sequences was identified as Raucaffricine-O-β-D-glucosidase (RG; EC 3.2.1.125). The two Rhazya sequences and RG were 76% and 77% identical whereas identities to Rauvolfia SGD were 58% and 57% at the amino acid level.
Thus it appears that the R. stricta genome may not encode SGD, nonetheless our own mass spectrometry analysis (Supplementary Table S9) along with decades of reports demonstrate the production of MIAs that derive from the reactive strictosidine aglycone, a product of SGD activity. The presence of a close homolog of RG, however, suggests another route to downstream intermediates and ultimately MIA end products. Raucaffricine, the major substrate of RG, is hydrolyzed to vomilenin, an intermediate in the ajmaline pathway11. While SGD is unable to hydrolyze raucaffricine, RG can catalyze the conversion of strictosidine12 and may fill that role in R. stricta cells. Identification and annotation of the SGD gene in a low quality nuclear genome draft from C. roseus proved challenging13, however, given the depth and completeness of our assembly it is unlikely that a Rhazya SGD sequence went undetected. Recently a similar approach used to search for MIA pathway genes in Amsonia hubrichtii (Apocynaceae, Rauvolfioideae) failed to detect a high identity homolog of the C. roseus SGD. A single transcript was designated as encoding a putative SGD with 57% amino acid identity to the Catharanthus sequence14, suggesting that another β-glucosidase may fill the role of SGD in this species.
The incredible diversity of MIAs in the Rauvolfioidae is facilitated through the hydrolysis of strictosidine to its aglycone, which may then be converted into different structural classes and elaborated by myriad reactions, many of which have been characterized, at least for C. roseus. However the enzymatic activities responsible for directing which structural class of MIA will ultimately derive from the aglycone moiety were largely unknown, until recently. THAS activity (Tetrahydroalstonine synthase) converts the strictosidine aglycone to tetrahydroalstonine, the gateway molecule to other heteroyohimbine type alkaloids such as cathenamine15,16. We queried our database with the THAS amino acid sequence, extracted homologs for gene phylogenies and produced trimmed, reconciled trees. Much like the other MIA pathway genes examined, THAS family members showed a pattern of lineage specific expansion in the Rauvolfioideae and included two sequences from R. stricta in the reduced tree (Fig. 3, Supplementary Fig. S2). While the two proximal sequences had only 60% amino acid identity to each other, their identities to the characterized C. roseus THAS was greater, at 71% and 75%.
Access to the high quality draft of the Coffea nuclear genome, along with drafts representing other lineages outside of Gentianales, has allowed us to use the Rhazya nuclear genome comparatively to evaluate lineage specific evolution of MIA pathway sequences. However, limiting these analyses to a single representative from each lineage necessarily limits the inferences that can be drawn from comparative study. The highly fragmented draft of the C. roseus genome13, while providing a useful resource for certain investigations, was not suitable for inclusion in the gene order comparisons. A more recent approach using single platform nuclear genome sequencing with PacBio generated a high quality draft for the grass genus Oropetum17. As sequencing technologies continue to improve in terms of both data quality and expense, less fragmented draft genomes for other Apocynaceae should become more tenable for future analyses.
Nonetheless, the gene phylogeny analyses facilitated by the Rhazya sequences combined with available data indicate lineage specific expansion resulting in distinct sets of MIA enzyme homologs in the Rauvolfioideae as well as in the Gentianales. The protein sequences clustering with characterized MIA homologs provide an evolutionary insight that could guide full characterization of MIA pathways in related species. One example may be found among the homologs of C. roseus Reticuline oxidase (RO, EC 1.21.3.3). This cytochrome P450 flavoprotein is a member of the BBE (Berberine bridge enzyme) family (pfam 08031) that catalyzes carbon-carbon bond formation via oxidative cyclization. An enzyme possessing such activity has been characterized in extracts from R. serpentina, but as yet the gene for SBE (Sarpagan bridge enzyme)18 remains unknown. The inclusion of several Rauvolfia and Rhazya sequences in the clade containing C. roseus RO (Supplementary Fig. S2m) may indicate an avenue for future studies seeking to identify elusive MIA pathway genes.
As more high quality nuclear genome drafts become available we will be able to address genome scale hypotheses of evolution. The rearrangements in Rhazya but not Coffea, relative to their shared ancestor may suggest that genome level dynamics played a role in the evolution of chemical diversity in the Rauvolfioideae. A critical evaluation of this question would require more than a single representative from each lineage to draw meaningful conclusions.
Materials and Methods
Plant material, DNA and RNA isolation
Rhazya stricta seeds were obtained from natural populations collected in the Makkah Province, Saudi Arabia. Seeds were soaked in water overnight at 37 °C then transferred to Profile® Field & Fairway™ inorganic ceramic particles (Buffalo Grove, IL) in a growth chamber (16 h light, 8 h dark 38 °C) for germination. Young leaves were flash frozen in liquid nitrogen for DNA and RNA isolation and stored at −80 °C. Isolation of R. stricta nucleic acids was described in Park et al.19. Young leaves of R. stricta were collected at the same location and shipped to INDRAS Private Limited (Hyderabad India) for mass spectrometry analysis of monoterpene indole alkaloid content.
For optical mapping, a megabase DNA isolation protocol was used to obtain intact nuclei from R. stricta for embedding in agarose. The protocol was downloaded from the Arizona Genomics Institute at the University of Arizona (http://www.genome.arizona.edu/modules/publisher/item.php?itemid=24).
Genome Sequencing
Illumina (San Diego, CA) DNA libraries were sequenced on the Illumina HiSeq 2500 at the Genome Sequencing and Analysis Facility at the University of Texas at Austin (GSAF). Three different library types were prepared, as required by the AllPaths-LG assembler (http://www.broadinstitute.org/software/allpaths-lg/blog/)20, an overlapping library with size range of 160–220 base pairs (bp), a mate-pair library ranging from 2–3 kilobases (kb) and a 6 kb mate-pair library. In addition, eight single molecule, real time (SMRT) cells of single-end PacBio RS reads were generated from a 10 kb library at the Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville. Supplementary Table S1 provides information about the data generated.
Transcriptome Sequencing
Total RNA isolation, library construction and Illumina sequencing were performed according to Zhang et al.21. Duplex specific nuclease normalization (Evrogen, Moscow, Russia) of the RNA samples, Illumina RNAseq library construction and sequencing were carried out at GSAF. Duplex specific nuclease normalization (Evrogen, Moscow, Russia) of the RNA samples, Illumina RNAseq library construction and sequencing were carried out at the GSAF. Raw read output from R. stricta RNAseq was deposited in the Small Read Archive (SRA) at the NCBI (accession number SRR1151604)18. The RNASeq data was assembled using Trinity (v2013_08_14)22. The assembly was annotated using Trinotate (https://trinotate.github.io). Trinotate collects annotation information into one database after running a number of annotation tools: homology search to SwissProt/Uniref90, protein domain identification using HMMER and PFAM, transmembrane domain prediction using tmHMM and comparison to functional categories such as Gene Ontology (GO) databases.
Genome assembly
A sequential pipeline of Illumina and PacBio tools were used to generate the assembly. An initial assembly was generated by Allpaths-LG20 (release 44837), using reads from three Illumina libraries (fragment library and overlapping 3 kb and 6 kb libraries; described above). Allpaths-LG default parameters were used with ploidy set to two. The assembly comprised 1449 scaffolds with scaffold N50 of 1.0 Mb. PacBio reads were added to the assembly for scaffolding using AHA scaffolder v1.2.0, part of SMRT Analysis 2.0 suite (http://www.pacb.com/products-and-services/analytical-software/devnet/devnet-analysis-tools/). The AHA scaffolder uses BLASR23 to map long PacBio reads to the assembly without gap filling. Blasr parameters required a minimum seed match of 10 bp (-minMatch 10), minimum identity of 70% (-minPctIdentity 10), reporting 10 best matches (-bestn 10); spliiting of subreads was not permitted (-noSplitSubreads). Parameters for the AHA hybrid assembly were varied iteratively and are provided in Supplementary Text S1. Further alignment of long PacBio reads with the draft assembly to close or improve gaps was carried out in PBJelly (v12.9.14; http://sourceforge.net/projects/pb-jelly/)24. The protocol file used to run all PBJelly stages (setup, mapping, support, extraction, assembly, output) is provided in Supplementary Text S1.
Rhazya nuclei embedded in agarose and the sequence of these 232 scaffolds were provided to OpGen (Gaithersburg, MD) for optical mapping. OpGen generated 11 high density Argus® MapCards using the BamHI restriction enzyme to obtain single molecule restriction maps for the Rhazya genome. Utilizing the initial R. stricta assembly and OpGen’s Genome Builder™ tool, OpGen compared the single molecule restriction data to the initial assembly to facilitate joining of scaffolds. If an overlap was detected when aligning the original Rhazya scaffolds to OpGen’s extension, OpGen assigned join scores. A join score of 9.0 or above indicated reliable joins. The 232 scaffolds were reduced to 113 scaffolds and the scaffold N50 increased from 1.45 Mb to 5.55 Mb. The whole genome map statistics provided by OpGen under a fee-for-service contract are available in Supplementary Table S3.
Genome annotation
The MAKER2 annotation pipeline (v2.28b; https://www.biostars.org/p/45281/)25 was employed for genome annotation. RepeatMasker (v3-3-0; http://www.repeatmasker.org)26 was used to mask all repeat classes from Repbase (http://www.girinst.org/repbase/) and low complexity sequences. The RepeatRunner webserver (http://www.yandell-lab.org/software/repeatrunner.html)27 was used to identify transposable elements based on the RepeatRunner protein database.
The Trinity-assembled transcripts were used to query the UniProt database (http://www.uniprot.org). Identified ESTs and protein sequences were aligned to the Rhazya genome using BLASTN and BLASTX (blast 2.2.27+)28, respectively. Exonerate (v3-3-0; http://www.ebi.ac.uk/about/vertebrate-genomics/software/exonerate)29 was used to realign sequences identified by BLAST around splice sites to more accurately detect gene models. Est2genome (http://emboss.sourceforge.net/apps/release/6.6/emboss/apps/est2genome.html) was used for polishing the EST hits and protein2genome, part of Exonerate, was used for polishing the protein hits.
SNAP ab initio gene predictor (v.2006-07-28; http://korflab.ucdavis.edu/software.html)30 was used to predict gene models. SNAP is an easy to use, easy to train tool that is supported by the MAKER2 pipeline. The program was trained by running MAKER2 twice. In the first run, gene models were produced exclusively from EST and protein homology evidence. These gene models were used to train SNAP and MAKER2 was rerun allowing for ab initio predictions using the newly trained gene predictor.
MAKER2 integrated all the EST alignments, protein alignments and ab initio predictions and updated information such as splice sites and UTR locations based on all the evidence to generate a filtered set of annotations. In addition to a final list of filtered and modified gene models, a list of rejected ab initio predictions were also generated for further detection of missing genes. All parameters used to run the various stages of MAKER2 annotation are provided in Supplementary Text S1
An additional 34,232 proteins were predicted by SNAP. These putative protein sequences were used to query a reference database consisting of annotated genes from Arabidopsis thaliana, Solanum lycopersicum (tomato), Vitis vinifera (grape), Coffea canephora (coffee), Catharanthus roseus, Rauvolfia serpentina and Theobroma cacao (>50% identity, evalue >0.000). The MAKER2 validated genes were also used to query the same reference database with the same identity and evalue requirements. If a SNAP predicted gene yielded a qualified hit to a gene in the reference database that had not been previously identified among the 17,041 validated genes, it was included in the final set of genes (4123 additional genes, Supplementary Table S5).
Verification of genome assembly and annotation
The assembled Rhazya genome was checked for completeness by using CEGMA 2.5 (Core Eukaryotic Genes Mapping Approach)8. CEGMA consists of a core set of eukaryotic genes that are highly conserved across eukaryotic taxa. The CEGMA genes were mapped to the Rhazya genome and a report indicating the completeness of the genome was generated.
Functional categorization of genome
All Rhazya transcripts were functionally annotated and were assigned gene names, functions, generic Gene Ontology (GO) terms and Enzyme codes (EC) by blasting to the UniProt protein database using BLAST settings (BLASTX, report 1 hit, maximum e-value 1E−6, percent identity greater than 40).
Metabolic pathway prediction
A metabolic pathway database was created for Rhazya using the PathoLogic component of Pathway Tools (v18.0; http://brg.ai.sri.com/ptools/)31. This program inferred metabolic pathways by comparing the annotated Rhazya genes to the MetaCyc reference pathway database (v.2.0; http://www.metacyc.org)32. MetaCyc contains experimentally elucidated, non-redundant metabolic pathways for multiple organisms and serves as a high quality reference for predicting pathways specific to an organism of interest. It also contains information about the enzymes, chemical compounds, reactions and genes related to these metabolic pathways.
The Pathologic Pathway Predictor algorithm consists of two phases. In the reactome inference phase, the predictor evaluates every annotated gene product in the genome and infers the reactions catalyzed by that gene product. The reactions catalyzed by a gene product are inferred from three information fields present in the input: the EC number, gene product name (enzyme name) and Gene Ontology (GO) terms. In the pathway inference phase, the tool infers the metabolic pathways present in the organism based on the set of catalyzed reactions. A pathway is inferred if it is not missing more than one reaction, or if a reaction that is unique to that pathway is present in the organism of interest. Nucleotide and amino acid sequence information is not considered by Pathologic in making these inferences. RhaCyc is available as a community resource at http://rhacyc.icmb.utexas.edu:1555/.
Structural evolution analysis
We used SynMap (with default parameters) in the CoGe platform (https://genomevolution.org/CoGe/; https://www.genomevolution.org/CoGe/SynMap.pl), to find syntenic blocks between Rhazya and Coffea (Rubiaceae, Gentianales). A whole genome triplication, discovered by Jaillon et al.10 while sequencing the Vitis vinifera genome, is broadly accepted to have occurred at the root of the core eudicots. This polyploidization, known as γ(gamma), produced a twenty-one chromosome genome from the seven chromosome ancestor. It is not too difficult to reconstruct the general structure of the immediate post-γ chromosomes since they differ from the nineteen-chromosome Vitis genome in only five or six large rearrangements. This baseline genome is the ultimate resource for deducing gene-order change in the core eudicots33.
Within each pair of genomes among Rhazya, Vitis and Coffea, SynMap synteny blocks together with overall gene similarity and Ks scores were used to establish thresholds for distinguishing duplicate gene pairs due to speciation (orthologs) which were retained for our analysis, from the older, weaker and sparser gene pairs due to the gamma triplication (“out-paralogs”), which were discarded. The coloring of the blocks was according to the 21 ancestral core eudicot chromosomes, as published by Zheng et al.33.
The circular display of homologies arrayed on the Rhazya scaffolds and Coffea/Vitis chromosomes were drawn with CIRCOS (http://circos.ca/)34.
The Bergeron et al.35 algorithm for double-cut-and-join (DCJ) was used to finding the shortest path between two genomes. Xu’s 2008 median solver for DCJ36 was employed to infer the Gentianales ancestor.
Gene family analysis
The subject database comprised protein sequences for six taxa. The species selected for inclusion in the MIA gene family analyses were required to meet certain criteria. The Apocynaceae taxa examined were required to produce MIAs, which restricted the sampling to the subfamily Rauvolfioideae37. The other three taxa included have complete genomes, which provided important information about isoforms so that only one isoform would be included in the gene tree analyses. In particular, coffee was selected because it is in the Gentianales, produces MIAs and its complete genome is available9. Tomato was included as an asterid outgroup to the Gentianales as its transcriptome and complete nuclear genome were available38. Likewise, Theobroma cacao, which produces non-MIA alkaloids, was included as a rosid outgroup to the asterid clade and had the required data available39. Protein sequences were downloaded for five species and combined with Rhazya sequences. Sequence data was accessed from the following sites: Theobroma cacao (rosid; http://www.cacaogenomedb.org/Tcacao_genome_v1.1), Solanum lycopersicum (non-Gentianales asterid; https://solgenomics.net/gb2/gbrowse/ITAG2.3_genomic/), Coffea canephora (Rubiaceae, Gentianales; http://coffee-genome.org), Catharanthus roseus and Rauvolfia serpentina (Apocynaceae, Gentianales; http://medicinalplantgenomics.msu.edu). Sequences for each of 21 characterized proteins involved in the MIA pathways of Catharanthus roseus (L.) and/or Rauvolfia serpentina (Supplementary Table S7) were used to query the database using BLAST (e-value 1e−3) and sequences with identities ≥ 35% were extracted. A single isoform was retained for each unique genetic locus and the sequences were aligned with the GenBank homolog (query) using MUSCLE40 as implemented in Geneious (www.biomatters.com). Sequences covering < 50% of the length of the GenBank homolog were removed. If the database sequence extended beyond the GenBank homolog sequence, the extra sequence was trimmed. Following removal of sequences, alignments were rerun with MUSCLE for maximum likelihood (ML) analysis with PhyML41 in Geneious. Muscle settings included eight iterations with sequences grouped by similarity using the kmer 6_6 for the first iteration and pctid_kimura distance for all subsequent iterations and the neighbor joining clustering method. ML analyses used the Le Gascuel substitution model and Subtree Pruning and Regrafting topology search options.
A species tree was drawn in FigTree (see top Fig. 2) and follows APG (http://www.mobot.org/MOBOT/Research/APweb/) relationships. For reconciliation of gene trees with the species tree eight datasets were selected representing early and late stage enzymes in the MIA pathways of C. roseus and R. serpentina. For these eight (bold in Supplementary Table 7), an iterative series of alignment and ML tree generation was used to define a well-supported clade that included the GenBank homolog. For each gene family, sequences for the clade of interest were extracted and aligned with MUSCLE and a preliminary unrooted binary phylogenetic tree was computed using PhyML version 20131022 (GTR + Γ(4) model). PhyML parameters were set to optimize branch length, tree topology and substitution rate. A hundred non-parametric bootstraps were performed to assess clade support in the resulting tree.
The initial trees were refined, rooted and internal nodes labeled as duplication or speciation nodes for reconciliation with the species tree. Weakly supported branches, i.e. branches with bootstrap values under a given threshold, were removed leading to an unrooted non-binary tree. Different thresholds for the bootstrap value (20, 30, 40, 50, 60, 70, 80 and 90) were tested. For each obtained tree, all nodes were successively considered as potential roots. Then for each rooted non-binary tree, all binary refinements minimizing the reconciliation cost, i.e. the inferred number of duplication and loss operations according to the species tree, were computed using ProfileNJ42. Each tree output by ProfileNJ is a rooted binary tree with each internal node labeled as a duplication (red square) or speciation (green circle) and each lost gene reported on the tree by grafting a new branch (gray dashed branches) and leaf labeled with abbreviated names of the genomes affected by the loss. This was the set of trees used as replicates for node bootstrapping.
Per-site likelihood for all trees was computed with RaxML43 version 8.1.3 and SH and AU tests were performed with CONSEL44. Then, manual filtering was done to retain only the best trees according to both the likelihood score and the reconciliation cost (Supplementary Table S8). A consensus tree was computed from all retained best trees using consense, part of the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html), with extended majority rule for identifying most frequent subtrees. Among the best trees, the one sharing the closest topology to the consensus tree was chosen. As branch length and bootstrap values were lost for branches contracted with ProfileNJ, they were recomputed for each tree. Finally, a node support was computed as the frequency of replicates having that node, i.e. with the same incoming edge and the same inferred event (duplication or speciation).
Data Availability
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession MEJB00000000. The version described in this paper is version MEJB01000000. All phylogenetic alignments are available upon request to TAR or RJK.
Additional Information
How to cite this article: Sabir, J. S.M. et al. The nuclear genome of Rhazya stricta and the evolution of alkaloid diversity in a medically relevant clade of Apocynaceae. Sci. Rep. 6, 33782; doi: 10.1038/srep33782 (2016).
References
Luca, V. D., Salim, V., Atsumi, S. M. & Yu, F. Mining the biodiversity of plants: A revolution in the making. Science 336, 1658–1661 (2012).
Chae, L., Kim, T., Nilo-Poyanco, R. & Rhee, S. Y. Genomic signatures of specialized metabolism in plants. Science 344, 510–513 (2014).
Szabó, L. F. Rigorous biogenetic network for a group of indole alkaloids derived from strictosidine. Molecules 13, 1875–1896 (2008).
Gilani, S. A., Kikuchi, A., Shinwari, Z. K., Khattak, Z. I. & Watanabe, K. N. Phytochemical, pharmacological and ethnobotanical studies of Rhazya stricta Decne. Phytother. Res. 21, 301–307 (2007).
Smith, G. N. Strictosidine: a key intermediate in the biogenesis of indole alkaloids. Chem. Commun. Lond. 912–914 (1968).
Sears, J. E. & Boger, D. L. Total synthesis of vinblastine, related natural products and key analogues and development of inspired methodology suitable for the systematic study of their structure–function properties. Acc. Chem. Res. 48, 653 (2015).
Unamaba, C. I. N., Nag, A. & Sharma R. K. Next generation sequencing technologies: The doorway to the unexplored genomics of non-model plants. Front. Plant Sci. 6, 1074 (2015).
Parra, G., Bradnam, K. & Korf, I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007).
Denoeud, F. et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345, 1181–1184 (2014).
Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007).
Ruppert, M., Panjikar, S., Barleben, L. & Stöckigt, J. Heterologous expression, purification, crystallization and preliminary X-ray analysis of raucaffricine glucosidase, a plant enzyme specifically involved in Rauvolfia alkaloid biosynthesis. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 62, 257–260 (2006).
Warzecha, H., Obitz, P. & Stöckigt, J. Purification, partial amino acid sequence and structure of the product of Raucaffricine-O-β-d-glucosidase from plant cell cultures of Rauwolfia serpentina. Phytochemistry 50, 1099–1109 (1999).
Kellner, F. et al. Genome-guided investigation of plant natural product biosynthesis. Plant J. 82, 680–692 (2015).
Xiao, M. et al. Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J. Biotechnol. 166, 122–134 (2013).
O’Connor, S. E. & Maresh, J. J. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat. Prod. Rep. 23, 532–547 (2006).
Stavrinides, A. et al. Unlocking the diversity of alkaloids in Catharanthus roseus: Nuclear localization suggests metabolic channeling in secondary metabolism. Chem. Biol. 22, 336–341 (2015).
VanBuren, R. et al. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 10.1038/nature15714 (2015).
Schmidt, D. & Stöckigt, J. Enzymatic formation of the sarpagan-bridge: A key step in the biosynthesis of sarpagine- and ajmaline-type alkaloids. Planta Med. 61, 254–258 (1995).
Park, S. et al. Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids. BMC Genomics 15, 405 (2014).
Ribeiro, F. et al. Finished bacterial genomes from shotgun sequence data. Genome Res. 22, 2270–7 (2012).
Zhang, J., Ruhlman, T. A., Mower, J. P. & Jansen, R. K. Comparative analyses of two Geraniaceae transcriptomes using next-generation sequencing. BMC Plant Biol. 13, 228 (2013).
Grabherr, M. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnol. 29, 644–652 (2011).
Chaisson, M. & Tesler, G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13, 238 (2012).
English A. C. et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7, e47768 (2012).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 (2011).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics 25, 4.10.1–4.10.14 (2009).
Smith, C. D. et al. Improved repeat identification and masking in dipterans. Gene 389, 1–9 (2007).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2008).
Slater, G. S. & Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 6, 31 (2005).
Korf, I. Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004).
Karp, P. D., Latendresse, M. & Caspi, R. The Pathway Tools pathway prediction algorithm. Stand Genomic Sci 5, 424–429 (2011).
Caspi, R. et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 44, D471–80 (2014).
Zheng, C., Chen, E., Albert, V. A., Lyons, E. & Sankoff, D. Ancient eudicot hexaploidy meets ancestral eurosid gene order. BMC Genomics 14, S3 (2013).
Krzywinski, M. et al. Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Bergeron, A., Mixtacki, J. & Stoye, J. In Algorithms in Bioinformatics (eds Bücher, P. & Moret, B. M. E. ) 4175, 163–173 (Springer Berlin Heidelberg, 2006).
Xu, A. W. & Sankoff, D. In Algorithms in Bioinformatics (eds Crandall, K. A. & Lagergren, J. ) 25–37 (Springer Berlin Heidelberg, 2008).
St-Pierre, B. et al. Deciphering the evolution, cell biology and regulation of monoterpene indole alkaloids. Adv. Bot. Res. 68, 73–109 (2013).
The Tomato Genome Consortium. The tomato genome sequence provides insights in to fleshy fruit evolution. Nature. 485, 635–641 (2012).
Argout, X. et al. The genome of Theobroma cacao. Nature Genet. 43, 101–108 (2011).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Guindon, S. & Gascuel, O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003).
Noutahi, E. et al. Efficient gene tree correction guided by species and synteny evolution. at https://hal.archives-ouvertes.fr/hal-01162963 (2015)
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Shimodaira, H. & Hasegawa, M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247 (2001).
Acknowledgements
The authors acknowledge financial support from Vice President for Educational Affairs Prof. Dr. Abdulrahman O. Alyoubi at King Abdulaziz University (KAU) and from the Deanship of Scientific Research represented by the Unit of Strategic Technologies Research at KAU through project Number 431/008-D. We thank the Texas Advanced Computing Center (TACC) for access to supercomputers, the Plant Resources Center for storage of voucher specimens and Scott Hunicke-Smith at the Genome Sequence and Analysis Facility (GSAF) at the University of Texas at Austin.
Author information
Authors and Affiliations
Contributions
J.S.M.S., R.K.J., T.A.R., D.A., M.A.K. and N.A.B designed and conceived research; T.A.R germinated seeds and harvested plants, isolated DNA, RNA and nuclei, assisted in data analyses and figure production and wrote the manuscript; D.A. assembled and annotated the genomic and transcriptomic data, performed blast comparisons for gene trees and developed RhaCyc; V.C., E.N. and N.E.-M. performed alignments and phylogenetic analyses on 8 selected MIA genes and gene tree/species tree reconciliation; S.P. assisted in transcriptome assembly and phylogenetic analyses; M.J.S., M.N.B., N.H.H., A.Y.O. and A.L.A.-M. acquired seed and leaf material of Rhazya stricta; C.Z. and D.S. performed gene order analyses; R.K.J. performed alignment and phylogenetic analyses of 21 MIA genes. All authors read and approved the final version of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Sabir, J., Jansen, R., Arasappan, D. et al. The nuclear genome of Rhazya stricta and the evolution of alkaloid diversity in a medically relevant clade of Apocynaceae. Sci Rep 6, 33782 (2016). https://doi.org/10.1038/srep33782
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep33782
This article is cited by
-
On the complexity of non-binary tree reconciliation with endosymbiotic gene transfer
Algorithms for Molecular Biology (2023)
-
Chromosome-level genome assembly of Ophiorrhiza pumila reveals the evolution of camptothecin biosynthesis
Nature Communications (2021)
-
Resolution effects in reconstructing ancestral genomes
BMC Genomics (2018)
-
Biotechnology of the medicinal plant Rhazya stricta: a little investigated member of the Apocynaceae family
Biotechnology Letters (2017)
-
Analysis of transcriptional response to heat stress in Rhazya stricta
BMC Plant Biology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.