Abstract
The sense of smell, or olfaction, is fundamental in the life of animals. However, penguins (Aves: Sphenisciformes) possess relatively small olfactory bulbs compared with most other waterbirds such as Procellariiformes and Gaviiformes. To test whether penguins have a reduced reliance on olfaction, we analyzed the draft genome sequences of the two penguins, which diverged at the origin of the order Sphenisciformes; we also examined six closely related species with available genomes and identified 29 one-to-one orthologous olfactory receptor genes (i.e. ORs) that are putatively functionally conserved and important across the eight birds. To survey the 29 one-to-one orthologous ORs in penguins and their relatives, we newly generated 34 sequences that are missing from the draft genomes. Through the analysis of totaling 378 OR sequences, we found that, of these functionally important ORs common to other waterbirds, penguins have a significantly greater percentage of OR pseudogenes than other waterbirds, suggesting a reduction of olfactory capability. The penguin-specific reduction of olfactory capability arose in the common ancestor of penguins between 23 and 60 Ma, which may have resulted from the aquatic specializations for underwater vision. Our study provides genetic evidence for a possible reduction of reliance on olfaction in penguins.
Similar content being viewed by others
Introduction
Traditionally, animals are believed to have five basic senses: sight, hearing, taste, touch and smell1. All the five sensory modalities are able to perceive stimuli from the external environment and are thus of fundamental importance for animals’ survival. However, one or more of the five senses could be absent in some species of animals2,3. Even when animals possess same sensory modalities, their reliance on each sense may vary significantly among species. For example, mice have an increased reliance on olfaction compared to humans4, while vampire bats showed decreased dependence on taste relative to other bats5,6.
Of the five basic senses, the sense of smell, or olfaction, is fundamentally important in the life of animals, underpinning many essential behaviors such as food location, mate recognition and predator avoidance7. Most mammals and reptiles typically possess two distinct olfactory systems: the main olfactory system (MOS) and the vomeronasal system (VNS), which were generally assumed to perceive environmental odorants and intraspecific pheromones, respectively8. The MOS was observed to primarily express olfactory receptors (ORs), which are encoded by olfactory receptor genes (ORs). The binding of odorants to ORs triggers the transduction of olfactory signals to the olfactory bulb in the brain, which results in olfactory perception9. The ORs make up one of the largest gene families in most vertebrate genomes, but the total number of ORs in each species varies dramatically, ranging from 125 in the pufferfish to 2129 in the cow10. The striking variation in the number of ORs may result from ecological adaptation. For instance, the loss of ORs occurred independently in the multiple lineages of aquatic mammals such as cetaceans and sirenians, which coincided with their habitat transition from land to water11,12. Furthermore, differences in the number of ORs may also reflect a tradeoff between olfaction and other senses. For instance, the reduction of ORs coincided with the independent acquisition of trichromatic color vision in multiple lineages of primates13. As such, investigating the evolutionary changes of ORs would provide a valuable route to understanding how genomes have been shaped by habitat transitions, sensory tradeoffs and other ecological adaptations.
Birds are the most species-rich group among tetrapod vertebrates, with diverse and distinct olfactory abilities. In general, they are assumed to have a poor olfactory system, as they seem to rely more on vision and vocalizations14. Indeed, birds were observed to have a much smaller OR gene repertoire than their closely related cousin, the reptiles15,16, which suggested a reduced reliance on olfaction. By contrast, numerous studies have argued that birds definitely use olfactory cues in many crucial behaviors, such as foraging, navigation and individual recognition17,18,19,20, which was also evidenced by recent genetic data21. The contrasting arguments with respect to avian olfaction call for further investigations.
As the only extant group of birds that occupy a secondarily aquatic niche with flightless wing-propelled diving, penguins (order Sphenisciformes) have undergone remarkable adaptations, such as streamlined bodies, flipper-like wings, dense bones and scale-like feathers22,23,24. In terms of sensory ecology, penguins are considered visual specialists, with a flat cornea and a spherical lens for underwater adaptation25,26. Consistent with the morphological adaptations, genetic studies have observed positive selection on phototransduction genes and accelerated evolution of visual opsin genes in penguins, which were also linked to the aquatic lifestyle27,28. By contrast, penguins have long been believed to lack a sense of smell, as they primarily rely on vision for underwater foraging22. In fact, multiple lines of evidence have demonstrated that penguins possess a functional sense of smell. For example, they smell the dimethyl sulphide (DMS) for prey location29,30,31 and perceive odors for kin recognition32. At the molecular level, penguins, along with other waterbirds (or aquatic birds), were identified to carry a significantly greater number of olfactory receptor genes, as compared with vocal-learning birds in a recent genomic analysis15, which also suggested that penguins rely heavily on olfaction. The recent genomic analysis classified the 48 birds into four categories: birds of prey, waterbirds, land birds and vocal learners and attempted to link the differences in OR gene numbers to ecological variations in the four categoires of birds15. However, the differences in OR gene numbers within one of the four categories were not examined15. Specifically, differences in olfaction between penguins and other waterbirds remain largely unknown. Compared with most other waterbirds, such as Procellariiformes, Ciconiiformes and Gaviiformes, penguins possess relatively small olfactory bulbs33, which are commonly used as a proxy for olfactory capability34. As such, we hypothesize that penguins may have a reduced number of OR genes as compared to other waterbirds. To test this hypothesis, we analyzed the draft genome sequences from the two penguins, which diverged at the origin of the order Sphenisciformes and represented the two major clades of the penguin species tree35; we also examined six closely related outgroup species with available genomes (Fig. 1) and identified 29 one-to-one orthologous olfactory receptor genes that are putatively functionally conserved and important across the eight birds. With additional sequencing of these orthologous genes, we found that penguins have a significantly higher percentage of pseudogenes than other waterbirds, although they still retain many intact and putatively functional genes.
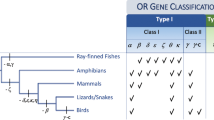
Genomic survey of olfactory receptor genes in the two penguins and six other waterbirds.
The eight birds represent four avian orders, their phylogeny follows a recent study36. The sequencing coverage of each genome was shown under each common name of birds. A filled square indicates a gene with a full-length coding sequence and an intact open reading frame, whereas an open square refers to either a partial gene or an absence of genomic data. Genes that are potentially lost in penguins were highlighted in red.
Results
Survey of OR genes in the genomes of penguins and other waterbirds
Our genomic dataset of birds represented four avian orders, including two species of Sphenisciformes (Emperor penguin, Aptenodytes forsteri, 60× coverage; Adelie penguin, Pygoscelis adeliae, 60×), one species of Procellariiformes (Northern fulmar, Fulmarus glacialis, 33×), four species of Pelecaniformes (Crested ibis, Nipponia Nippon, 105×; Little egret, Egretta garzetta, 74×; Great cormorant, Phalacrocorax carbo, 24×; Dalmatian pelican, Pelecanus crispus, 34×) and one species of Gaviiformes (Red-throated loon, Gavia stellata, 33×) (Fig. 1; Supplementary Table S1)36,37. The four orders of birds are closely related and were referred to as the core waterbirds36. Specifically, the penguin order (Sphenisciformes) is the most closely related to the order Procellariiformes; the two orders form a monophyletic group, which is clustered with the order Pelecaniformes; and the fourth order Gaviiformes falls outside of the other three orders (Fig. 1)36. In addition, the two penguins (i.e. the emperor penguin and the Adelie penguin) with genome sequences diverged at the origin of the order Sphenisciformes and represented the two major clades of the penguin species tree35.
A total of 344 full-length and intact ORs (see the identification procedure in Materials and Methods) were identified from the draft genome sequences of the emperor penguin (gene number: 32), Adelie penguin (26), northern fulmar (33), great cormorant (36), crested ibis (47), little egret (106), Dalmatian pelican (20) and red-throated loon (44) (Supplementary Table S1). The numbers of identified intact ORs in the present study are similar to those from a recent study with minor differences (Supplementary Table S1), which may result from slightly different bioinformatics approaches between the current work and a previous study15. Relative to most mammals, the low number of intact ORs in birds suggests a reduced reliance on olfaction, which is consistent with the common view that most birds are primarily visual animals38, because the reduction of ORs was coincident with the occurrence of better color vision in primates13. Notably, the little egret has an extraordinarily large number of ORs, indicating an extraordinary expansion as compared to other waterbirds15.
Phylogenetic analysis was performed using all intact ORs from the eight avian genomes. The resulting phylogenetic tree revealed a major clade comprising mostly the little egret genes, although the clade was not well supported by both phylogenetic methods (Supplementary Fig. S1). This clade consisted of 94 genes in which 75 genes are from the little egret, whereas the remaining clades contain only 31 little egret genes, suggesting an apparent gene expansion in the little egret (Supplementary Fig. S1), which was also observed in a recent analysis15. Of note, the monophyly of the major clade remains to be resolved in future. While the basal clades of the phylogenetic trees did not receive high supporting values, well-supported clades were found at the tips of many groupings (Supplementary Fig. S1). We identified 29 potentially one-to-one orthologous genes with the following criteria. First, we selected well-supported clades with a bootstrap value greater than 85%; Second, there is only one single-copy gene from each species; Third, the single-copy gene was detected from at least four avian species in a well-supported clade. The nomenclature of the 29 genes followed the HORDE database (Supplementary Fig. S1)39. For convenience, we also named each of the 29 genes numerically in the order of appearance on the phylogenetic tree (Supplementary Fig. S1). These one-to-one orthologous genes are assumed to be functionally conserved in the eight birds, because the same olfactory receptors tend to detect similar odorants40. As such, the 29 orthologous genes are expected to be present in most of the eight avian species and some genes are absent possibly because of incomplete genome sequencing or poor genome assembly.
To test whether penguins have specifically lost some of the 29 orthologous genes, we mapped these genes onto the species phylogeny of the two penguins and six related waterbirds36. Although 163 ORs were detected, a total of 69 ORs were not identified with a full-length and intact ORF (open reading frame) from the draft genomes of the emperor penguin (number of unidentifiable genes: 11), Adelie penguin (15), northern fulmar (9), great cormorant (12), crested ibis (1), little egret (7), Dalmatian pelican (6) and red-throated loon (8) (Fig. 1). It is understandable that the crested ibis has just 1 of the 29 OR genes that cannot be identified from its genome, because its genome coverage is the highest (105×). Similarly, the large number (12) of unidentifiable genes in the great cormorant could be attributed to its having the lowest genome coverage (24×) (Fig. 1). However, there are 11 and 15 unidentifiable genes in the emperor penguin and Adelie penguin with high-coverage genomes, respectively and the numbers of unidentifiable genes in both penguins are even greater than those in three other waterbirds (the northern fulmar, red-throated loon and Dalmatian pelican) with low-coverage genomes (Fig. 1). This finding suggests that penguins may have lost more functionally conserved ORs than other waterbirds. In particular, 11 ORs (OR3-4, OR7, OR9, OR13, OR19, OR22, OR24-27) were not identified in both penguins (Fig. 1), indicating penguin-specific losses.
Pseudogenization of OR genes in penguins
To examine whether some orthologous ORs were specifically lost in penguin lineages, we attempted to sequence all missing data (Fig. 1). In case of missing data from the two penguin genomes, we performed gene sequencing using the genomic DNAs of their respective congeners (King penguin and Chinstrap penguin); the northern fulmar, red-throated loon and little egret were examined with PCRs; the crested ibis was also included for comparison without additional sequencing, because this bird only missed one OR gene from its genome, which appeared to be intact in penguins (Fig. 2). We did not examine the great cormorant and Dalmatian pelican due to the absence of genetic material, although this dataset of six birds still represents the four avian orders36. Of note, we did not sequence the two species of penguins with available genomes due to the lack of genomic DNAs, but their congeneric species are still appropriate to infer pseudogenization events in the common ancestor of all penguins by identifying shared frameshifts or interrupting stop codons.
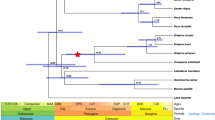
Survey of avian olfactory receptor genes by additional sequencing.
In case that OR gene sequences are missing from the two penguin genomes, genomic DNAs of their congeneric species (King penguin and Chinstrap penguin) were used to perform amplification and sequencing. The great cormorant and Dalmatian pelican were not included because of the lack of genetic material, but the examined species still represent the four avian orders. Genes that are potentially lost in penguins were highlighted in red.
We sequenced 34 OR gene segments from the five birds as mentioned earlier (Fig. 2), which ranged from 612 to 964 base pair (bp), with an average of 805 bp (National Center for Biotechnology Information accession numbers: KX171590-KX171621 and KX189196-KX189197). Phylogenetic analysis was used to confirm the orthology of each gene. The newly acquired 34 sequences were aligned with 121 ORs identified from draft genomes, which resulted in a total of 155 OR sequences for subsequent analysis. We found 134 out of 155 ORs to have intact open reading frames (ORFs) (Fig. 2). Among the four non-penguin birds (the northern fulmar, crested ibis, little egret and red-throated loon), 99 out of 102 ORs (99/102 = 97.1%) were identified with intact ORFs (Fig. 2), supporting our assumption that these genes are functionally important across non-penguin waterbirds. By contrast, 36 out of 54 ORs (36/54 = 66.7%) were found to be intact in penguins (Fig. 2). Among those pseudogenized ORs containing ORF-disrupting mutations such as nonsense mutations and frame-shifting insertions or deletions, 18 were found in penguins; the remaining 3 genes are from non-penguin birds (Fig. 2). In 17 out of the 21 pseudogenized ORs, the first nonsense mutations resulted from ORF-disrupting mutations are located near 5′ end of each gene (Fig. 3), which would lead to the loss of multiple transmembrane domains of each protein. The 3 pseudogenes (OR7 of the king penguin, OR7 of the chinstrap penguin and OR9 of the king penguin) contained the first nonsense mutations located near the 3′ end of each gene (Fig. 3 and Supplementary Fig. S2), which could also result in the loss of the final transmembrane domain of an olfactory receptor41. We did not observe a nonsense mutation in the pseudogenized OR9 of the chinstrap penguin, but we observed a 2-bp deletion shared by both penguins (Fig. 3). This finding suggested that none of the 21 pseudogenes could encode a functional olfactory receptor. Therefore, after examining these functionally important ORs common to non-penguin waterbirds, we found that the percentage of nonfunctional ORs is significantly greater in penguins (18/54 = 33.3%) than in their closely related non-penguin waterbirds (3/102 = 2.9%) (p < 0.0001, Fisher’s exact test), which suggested a penguin-specific reduction of olfactory capability.
Nucleotide alignments of avian olfactory receptor genes.
The first ORF-disrupting mutations and the followed common mutations were boxed. Dashes indicate alignment gaps and numbers in parentheses represent nucleotide positions following the reference sequences from either Northerm fulmar or Crested ibis.
Among the 29 orthologous ORs, 8 were sequenced from both penguins (the king penguin and chinstrap penguin) (Figs 2 and 3). With an exception of OR19, at least one common ORF-disrupting mutation was identified between the two penguins for each orthologous OR gene. For example, OR3 contained one 2-bp deletion and one 2-bp insertion that are shared between the two penguins; OR4 included one shared nonsense mutation; OR7 has one shared 10-bp deletion and two common nonsense mutations (Fig. 3 and Supplementary Fig. S2). We did not observe any shared ORF-disrupting mutations in OR19, but we identified a relatively large deletion (12-bp) that are common in the two penguins; coupled with multiple nonsense mutations ahead of the 12-bp deletion, this finding suggested that OR19 was pseudogenized prior to the divergence of the two penguins (Fig. 3). Given that the king penguin and the chinstrap penguin diverged at the origin of the order Sphenisciformes, our genetic evidence strongly suggests that at least 8 functionally important ORs in other waterbirds were lost in all penguins and the relaxation of functional constraints on these olfactory receptor genes occurred predating the divergence of penguins. Because penguins diverged 23 million years ago (Ma) and penguins diverged from their closest relatives (order Procellariiformes) approximately 60 Ma28,36, the penguin-specific reduction of olfactory capability took place in the common ancestor of penguins between 23 and 60 Ma. In addition, we observed three independent pseudogenizations in non-penguin waterbirds (OR10 and OR16 in the northern fulmar, OR25 in the red-throated loon) (Supplementary Fig. S2).
To understand why penguins could afford to lose some important ORs that are common in other waterbirds, we assigned each pseudogenized OR into a specific OR gene family following a recent study42. Among the 29 putative one-to-one orthologous ORs (OR1-OR29), 8 genes (OR3, OR4, OR7, OR9, OR13, OR19, OR22 and OR25) were found to have common disruptive mutations between the two penguins, suggesting an ancestral pseudogenization in the common ancestor of penguins; 2 genes (OR24 and OR26) were sequenced in one of the two penguins because of the failure of PCRs; 1 gene (OR27) was not able to be amplified in both penguins even after trying several primer pairs (Figs 2 and 3). The failure of amplification suggests either severe degeneration or loss of these genes, we thus infer that 11 out of 29 ORs were pseudogenized in penguins. After using the OR family Assigner42, four genes (OR3, OR4, OR7 and OR9), three genes (OR13, OR19 and OR22), two genes (OR24 and OR25) and the remaining two genes (OR26 and OR27) were assigned into the OR gene family 52, family 5, family 10 and family 6, respectively. According to the traditional classification based on sequence similarity, ORs are divided into 18 families. Class I families (i.e. OR gene family 51–56) are assumed to detect water-borne molecules, whereas Class II families (i.e. OR gene family 1–14) are believed to recognize air-borne compounds42,43,44. Consequently, penguins have pseudogenized 4 ORs that could function underwater and 7 ORs that could smell in the air. In addition, an earlier study suggested that OR gene family 5 was associated with the foraging behavior of predatory birds and that OR gene families 6 and 10 were prominent in vocal-learning birds15. Therefore, penguins have lost several ORs that are important in vocal learner and birds of prey.
Discussion
Through examination of 8 draft genomes of 2 penguins and 6 non-penguin waterbirds that are closely related to penguins, we identified 29 ORs that are putative one-to-one orthologs among all 8 waterbirds by phylogenetic analysis. We next attempted to survey the functionality of the 29 ORs by sequence analysis in penguins and their relatives. With the aid of additional sequencing, we found that, of these functionally important ORs common to other waterbirds, penguins were found to have a significantly greater percentage of pseudogenized ORs than other waterbirds, suggesting a major reduction of olfactory capability in penguins relative to other waterbirds. However, the small number of olfactory receptor genes does not necessarily represent a reduced importance of olfaction in penguins, because olfactory receptors could evolve to recognize more odorants and develop novel receptors, as reported in taste receptors45,46. Despite this, our genetic evidence suggests that penguins appear to have a less developed sense of smell than most other waterbirds, as a higher number of chemosensory receptor genes allows the evolution of more specialized receptors45. Since genetic evidence is indirect, our study awaits a behavioral test to verify the possible reduction of olfaction in penguins.
In support of our genetic evidence, anatomical reduction of the olfactory bulb in penguins was observed as compared to most other waterbirds, which suggested that penguins generally have a reduction of olfactory acuity relative to other waterbirds33,34,47. Fossil evidence revealed that ancient penguins had much larger olfactory bulbs than extant penguins, suggesting that the reduction of olfactory acuity started shortly before the divergence of penguins48. However, the reduced olfaction does not suggest that penguins do not use the sense of smell. Behavioral studies have convincingly demonstrated that penguins can use olfaction to locate prey and recognize kins29,30,32. Indeed, our genetic analysis also revealed that penguins still retain many intact ORs, suggesting a functional role of olfaction in these birds. In fact, penguins were found to possess even more ORs than vocal-learning birds, suggesting that they have a better sense of smell than some other birds15.
Why could penguins afford to reduce the reliance on olfaction compared to other waterbirds? Penguins are the only surviving birds that inhabit a secondarily aquatic environment with flightless wing-propelled diving behavior; their reduced reliance on olfaction parallels the reduction in marine mammals that independently occupy a secondarily aquatic niche11,12,48. Indeed, we found penguins to have lost several OR genes that encode olfactory receptors detecting air-borne molecules, suggesting that penguins do not need to smell some air-borne molecules in the aquatic environment. Within mammals, sensory tradeoffs were proposed49,50; the reduction of OR genes in trichromatic rather than dichromatic primates has been explained by a trade-off between vision and olfaction13. However, this explanation cannot be the case in penguins, because penguins are trichromatic while most other birds are tetrachromatic27,28. On the other hand, positive selection on phototransduction genes and accelerated evolution of visual opsin genes were detected in penguins, which were associated with aquatic adaptation28. Furthermore, the photic adaptation in penguins has also been detected by morphological specializations such as flat corneas and spherical lens25,26. In addition, unlike other diving waterbirds, penguins spend the entire search, chase and capture underwater, some species of penguins are even able to dive 200 meters in depth and 30 minutes in duration51. Therefore, the aquatic specializations for underwater vision in penguins may have rendered their olfaction less important. Other ecological traits may also account for the reduction of olfactory acuity in penguins. For example, since penguins originated in the coldest niche on Earth, the extremely cold temperature of the Antarctic may have influenced the evolution of olfactory perception52, a hypothesis awaiting future empirical investigation. Taken together, we found genetic evidence for a possible reduction of reliance on olfaction in penguins and highlighted the power and necessity of in-depth genetic analysis based on draft genome sequences. Although the loss of three primary tastes in penguins has already been revealed53, future studies of other sensory systems in penguins and other waterbirds would provide a better understanding of how penguins could sense and survive in their unique ecological niche.
Materials and Methods
Genome data and gene identification
The draft genome sequences of the eight species of waterbirds were retrieved from the Avian Phylogenomics Project ( http://avian.genomics.cn/en/, last accessed March 25, 2015). Vertebrate ORs are single-exon genes that encode seven-helix transmembrane proteins40. To identify full-length and intact ORs from the eight waterbird genomes, we followed a standard protocol as described previously54. Briefly, we used full-length OR protein sequences55,56 as queries to conduct TBLASTN searches57 with an e-value cutoff of 1e-5. The best hits were determined with the criteria of the lowest e-value and the longest alignment and the putative start and stop codons were identified by extending in both 5′ and 3′ directions. All these potential OR genes were then compared (BLASTX) back to the NCBI non-redundant database and those with the best blast hit of a non-OR gene were discarded. Sequences that are longer than 250 amino acids and have no interrupting stop codons or frameshifts were aligned to known OR genes and those with a gap of five or more amino acids within transmembrane domains or other conserved regions were excluded. The remaining sequences were considered to be the full-length and intact OR genes, which are used for further analysis.
Phylogenetic reconstruction and OR gene family assignment
Phylogenetic reconstruction was conducted to identify putative one-to-one orthologous genes across the eight birds with draft genomes. A total of 344 complete and intact ORs identified from the eight avian genomes were analyzed with a zebrafish OR gene (GenBank: NM_001083869) as the outgroup. The 345 full-length ORs were translated into protein sequences in MEGA version 658 and were next aligned by MUSCLE59 with manual adjustments. The protein sequence alignment was subsequently translated back to nucleotide sequence alignment, which was used to reconstruct phylogenetic trees. Phylogenetic analyses were performed with both Maximum Likelihood (ML) and Bayesian Inference (BI) approaches. The jModelTest2 program60 was used to infer the best-fitting substitution model and the model GTR + I + G was selected. The RAxML version 7.2.661 was used to reconstruct ML trees with bootstrap replicates of 1000. The Bayesian tree was constructed by MrBayes version 3.262 with six Markov chains and six million generations.
Genes identified from the whole genomes were assigned into OR gene families using the OR family Assigner, ORA version 1.942. Specifically, we undertook the domain-based hmmscan searches against the HMM (hidden Markov models) database using the program HMMER version 3.1b163. We assigned a given OR gene into an OR gene family with the lowest e-value produced from the hmmscan searches. The nomenclature of each OR gene (Supplementary data set S1) followed the best hit after conducting BLASTN searches against the HORDE database39. For convenience, we also named each gene with the order in which they were identified (Supplementary data set S1).
Taxon sampling and DNA sequencing
Due to the incomplete genome sequencing or poor genome assembly, some putative one-to-one orthologous genes were not identified with a complete and intact ORF (open reading frame) from the draft genomes (Fig. 1). To obtain the missing data, we designed new primers (Supplmentary Table S2) to resequence all missing sequences based on the nucleotide sequence alignments of each gene. The two penguins (i.e. the emperor penguin and the Adelie penguin) with genome sequences are from the two major clades of penguin species tree35, the ancestor of both penguins thus represents the common ancestor of all extant penguins. In case that OR gene sequences are missing from the penguin genomes, we attempted to amplify from the genomic DNAs of their congeneric species (King penguin, Aptenodytes patagonicus; Chinstrap penguin, Pygoscelis antarctica) (Fig. 2), which were left from a previous project53. The genomic DNAs of the northern fulmar and red-throated loon were from the same project in which we loaned all avian samples from the University of Michigan Museum of Zoology53. The muscle tissue of the little egret (Sample ID: IOZ-9748) was obtained from the Institute of Zoology, Chinese Academy of Sciences. We also included the crest ibis in our resequencing dataset but did not conduct the sequencing, because this bird only missed one OR gene from its genome, which appeared to be intact in penguins (Fig. 2). As a result, our dataset of resequencing contained six avian species: the king penguin, chinstrap penguin, northern fulmar, red-throated loon, little egret and crested ibis (Fig. 2), which can still represent the four orders (Sphenisciformes, Procellariiformes, Pelecaniformes and Gaviiformes)36.
Genomic DNA of the little egret was isolated with Qiagen DNeasy kit. Polymerase chain reactions (PCRs) and DNA sequencing were conducted according to procedures previously described3,5,6. Newly generated sequences by PCRs were aligned with their respective orthologous OR genes identified from the whole genomes. The phylogenetic tree of each gene was inferred to confirm the orthology relationship using a Bayesian approach as implemented in MrBayes version 3.2 62. Pseudogenes were identified when nonsense and/or frame-shifting mutations were observed in an OR sequence. All new sequences have been submitted to GenBank under accession numbers KX171590-KX171621 and KX189196-KX189197.
Additional Information
Accession Codes: DNA sequences: Genbank accessions KX171590-KX171621 and KX189196-KX189197.
How to cite this article: Lu, Q. et al. Penguins reduced olfactory receptor genes common to other waterbirds. Sci. Rep. 6, 31671; doi: 10.1038/srep31671 (2016).
References
Sorabji, R. Aristotle on demarcating the five senses. Phil. Review 80, 55–79 (1971).
Haim, A., Heth, G., Pratt, H. & Nevo, E. Photoperiodic effects onthermoregulation in a “blind” subterranean mammal. J. Exp. Biol. 107, 59–64 (1983).
Feng, P., Zheng, J. S., Rossiter, S. J., Wang, D. & Zhao, H. Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol Evol 6, 1254–1265 (2014).
Young, J. M. et al. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 11, 535-546 (2002).
Hong, W. & Zhao, H. Vampire bats exhibit evolutionary reduction of bitter taste receptor genes common to other bats. Proc Biol Sci 281, 20141079 (2014).
Zhao, H. et al. Evolution of the sweet taste receptor gene Tas1r2 in bats. Mol. Biol. Evol. 27, 2642–2650 (2010).
Doty, R. L. Mammalian olfaction, reproductive processes and behavior . (Academic Press, 1976).
Grus, W. E. & Zhang, J. Distinct evolutionary patterns between chemoreceptors of 2 vertebrate olfactory systems and the differential tuning hypothesis. Mol. Biol. Evol. 25, 1593–1601 (2008).
Ronnett, G. V. & Moon, C. G proteins and olfactory signal transduction. Annu Rev Physiol 64, 189–222 (2002).
Nei, M., Niimura, Y. & Nozawa, M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet 9, 951–963 (2008).
Hayden, S. et al. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res 20, 1–9 (2010).
Kishida, T., Kubota, S., Shirayama, Y. & Fukami, H. The olfactory receptor gene repertoires in secondary-adapted marine vertebrates: evidence for reduction of the functional proportions in cetaceans. Biol Lett 3, 428–430 (2007).
Gilad, Y., Wiebel, V., Przeworski, M., Lancet, D. & Paabo, S. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. Plos Biol. 2, 120–125 (2004).
Roper, T. J. Olfaction in birds. Adv. Study Behav. 28, 247–332 (1999).
Khan, I. et al. Olfactory receptor subgenomes linked with broad ecological adaptations in Sauropsida. Mol. Biol. Evol. 32, 2832–2843 (2015).
Steiger, S. S., Kuryshev, V. Y., Stensmyr, M. C., Kempenaers, B. & Mueller, J. C. A comparison of reptilian and avian olfactory receptor gene repertoires: species-specific expansion of group gamma genes in birds. BMC Genomics 10, 446 (2009).
Bonadonna, F. & Nevitt, G. A. Partner-specific odor recognition in an Antarctic seabird. Science 306, 835 (2004).
van Buskirk, R. W. & Nevitt, G. A. The influence of developmental environment on the evolution of olfactory foraging behaviour in procellariiform seabirds. J. Evol. Biol. 21, 67–76 (2008).
Wallraff, H. G. Navigation by homing pigeons: updated perspective. Ethol Ecol Evol 13, 1–48 (2001).
Caspers, B. A. & Krause, E. T. Odour-based natal nest recognition in the zebra finch (Taeniopygia guttata), a colony-breeding songbird. Biol Lett 7, 184–186 (2011).
Steiger, S. S., Fidler, A. E., Valcu, M. & Kempenaers, B. Avian olfactory receptor gene repertoires: evidence for a well-developed sense of smell in birds? Proc Biol Sci 275, 2309–2317 (2008).
Ksepka, D. T. & Ando, T. In Living dinosaurs (eds Dyke, G. & Kaiser, G. ) 155–186 (Wiley, 2011).
Taylor, J. R. E. Thermal insulation of the down and feathers of pygoscelid penguin chicks and the unique properties of penguin feathers. Auk 103, 160–168 (1986).
Meister, W. Histological structure of the long bones of penguins. Anat Rec 143, 377–387 (1962).
Sivak, J., Howland, H. C. & McGill-Harelstad, P. Vision of the Humboldt penguin (Spheniscus humboldti) in air and water. Proc Biol Sci 229, 467–472 (1987).
Sivak, J. G. The role of a flat cornea in the amphibious behaviour of the blackfoot penguin (Spheniscus demersus). Can J Zool 54, 1341–1345 (1976).
Borges, R. et al. Gene loss, adaptive evolution and the co-evolution of plumage coloration genes with opsins in birds. BMC Genomics 16, 751 (2015).
Li, C. et al. Two Antarctic penguin genomes reveal insights into their evolutionary history and molecular changes related to the Antarctic environment. GigaScience 3, 27 (2014).
Wright, K. L. B., Pichegru, L. & Ryan, P. G. Penguins are attracted to dimethyl sulphide at sea. J. Exp. Biol. 214, 2509–2511 (2011).
Cunningham, G. B., Strauss, V. & Ryan, P. G. African penguins (Spheniscus demersus) can detect dimethyl sulphide, a prey-related odour. J. Exp. Biol. 211, 3123–3127 (2008).
Cunningham, G. B., Leclaire, S., Toscani, C. & Bonadonna, F. Responses of King penguin (Aptenodytes patagonicus) adults and chicks to two food-related odours. J. Avian Biol., Forthcoming (2016).
Coffin, H. R., Watters, J. V. & Mateo, J. M. Odor-based recognition of familiar and related conspecifics: a first test conducted on captive Humboldt penguins (Spheniscus humboldti). Plos One 6, e25002 (2011).
Bang, B. G. & Cobb, S. The size of the olfactory bulb in 108 species of birds. Auk 85, 55–61 (1968).
Zelenitsky, D. K., Therrien, F., Ridgely, R. C., McGee, A. R. & Witmer, L. M. Evolution of olfaction in non-avian theropod dinosaurs and birds. Proc Biol Sci 278, 3625–3634 (2011).
Baker, A. J., Pereira, S. L., Haddrath, O. P. & Edge, K. A. Multiple gene evidence for expansion of extant penguins out of Antarctica due to global cooling. Proc Biol Sci 273, 11–17 (2006).
Jarvis, E. D. et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1320–1331 (2014).
Zhang, G. et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320 (2014).
Hart, N. S. The visual ecology of avian photoreceptors. Prog. Retin. Eye Res. 20, 675–703 (2001).
Olender, T., Feldmesser, E., Atarot, T., Eisenstein, M. & Lancet, D. The olfactory receptor universe--from whole genome analysis to structure and evolution. Genet Mol Res. 3, 545–553 (2004).
Gaillard, I., Rouquier, S. & Giorgi, D. Olfactory receptors. Cell. Mol. Life Sci. 61, 456–469 (2004).
Mombaerts, P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci 5, 263–278 (2004).
Hayden, S. et al. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 20, 1–9 (2009).
Glusman, G. et al. The olfactory receptor gene superfamily: data mining, classification and nomenclature. Mamm. Genome 11, 1016–1023 (2000).
Warren, W. C. et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453, 175–183 (2008).
Behrens, M., Korsching, S. I. & Meyerhof, W. Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes. Mol. Biol. Evol. 31, 3216–3227 (2014).
Baldwin, M. W. et al. Evolution of sweet taste perception in hummingbirds by transformation of the ancestral umami receptor. Science 345, 929–933 (2014).
Bang, B. G. Functional anatomy of the olfactory system in 23 orders of birds. Acta Anat. (Basel). 79, 1–76 (1971).
Tambussi, C. P., Degrangeab, F. J. & Ksepkacd, D. T. Endocranial anatomy of Antarctic Eocene stem penguins: implications for sensory system evolution in Sphenisciformes (Aves). J. Vert. Paleontol. 35, e981635 (2015).
Zhao, H. et al. The evolution of color vision in nocturnal mammals. Proc Natl Acad Sci USA 106, 8980–8985 (2009).
Zhang, J. & Webb, D. M. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc Natl Acad Sci USA 100, 8337–8341 (2003).
Cronin, T. W. Visual optics: Accommodation in a splash. Curr. Biol. 22, R871–R873 (2012).
Riveron, J., Boto, T. & Alcorta, E. The effect of environmental temperature on olfactory perception in Drosophila melanogaster. J. Insect Physiol. 55, 943–951 (2009).
Zhao, H., Li, J. & Zhang, J. Molecular evidence for the loss of three basic tastes in penguins. Curr. Biol. 25, R141–R142 (2015).
Niimura, Y. Identification of chemosensory receptor genes from vertebrate genomes. Methods Mol Biol 1068, 95–105 (2013).
Niimura, Y. & Nei, M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE 2, e708 (2007).
Niimura, Y. & Nei, M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci USA 102, 6039–6044 (2005).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J Mol Biol 215, 403–410 (1990).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 (2004).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9, 772 (2012).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Eddy, S. R. Profile hidden Markov models. Bioinformatics 14, 755–763 (1998).
Acknowledgements
The authors thank the University of Michigan Museum of Zoology for genetic material. We also thank Feng-Juan Mu for technical assistance in the laboratory. This work was supported in part by the National Natural Science Foundation of China (31300313).
Author information
Authors and Affiliations
Contributions
Q.L. carried out the molecular lab work, participated in data analysis and the design of the study; K.W. helped with data analysis; F.L. helped with sample collection and commented the manuscript; D.Y. commented the manuscript; H.Z. conceived of the study, designed the study, coordinated the study and wrote the manuscript. All authors gave final approval for publication.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Lu, Q., Wang, K., Lei, F. et al. Penguins reduced olfactory receptor genes common to other waterbirds. Sci Rep 6, 31671 (2016). https://doi.org/10.1038/srep31671
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep31671
This article is cited by
-
Signature of adaptive evolution in olfactory receptor genes in Cory’s Shearwater supports molecular basis for smell in procellariiform seabirds
Scientific Reports (2020)
-
Convergent reduction of V1R genes in subterranean rodents
BMC Evolutionary Biology (2019)
-
Exploration during early life: distribution, habitat and orientation preferences in juvenile king penguins
Movement Ecology (2019)
-
Transcriptome sequencing and phylogenetic analysis of four species of luminescent beetles
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.