Abstract
Dom34-Hbs1 stimulates degradation of aberrant mRNAs lacking termination codons by dissociating ribosomes stalled at the 3′ ends and plays crucial roles in Nonstop Decay (NSD) and No-Go Decay (NGD). In the dom34Δ mutant, nonstop mRNA is degraded by sequential endonucleolytic cleavages induced by a stalled ribosome at the 3′ end. Here, we report that ribosome-associated Asc1/RACK1 is required for the endonucleolytic cleavage of nonstop mRNA by stalled ribosome at the 3′ end of mRNA in dom34Δ mutant cells. Asc1/RACK1 facilitates degradation of truncated GFP-Rz mRNA in the absence of Dom34 and exosome-dependent decay. Asc1/RACK1 is required for the sequential endonucleolytic cleavages by the stalled ribosome in the dom34Δ mutant, depending on its ribosome-binding activity. The levels of peptidyl-tRNA derived from nonstop mRNA were elevated in dom34Δasc1Δ mutant cells and overproduction of nonstop mRNA inhibited growth of mutant cells. E3 ubiquitin ligase Ltn1 degrades the arrest products from truncated GFP-Rz mRNA in dom34Δ and dom34Δasc1Δ mutant cells and Asc1/RACK1 represses the levels of substrates for Ltn1-dependent degradation. These indicate that ribosome-associated Asc1/RACK1 facilitates endonucleolytic cleavage of nonstop mRNA by stalled ribosomes and represses the levels of aberrant products even in the absence of Dom34. We propose that Asc1/RACK1 acts as a fail-safe in quality control for nonstop mRNA.
Similar content being viewed by others
Introduction
Systems for quality control of aberrant mRNAs prevent production of potentially harmful protein products by repressing translation and promoting protein degradation and stimulating mRNA degradation1,2,3,4,5. No-Go Decay (NGD) of mRNA, which is triggered by blockage of translation during the elongation step, induces the endonucleolytic cleavage in the vicinity of the stalled site6,7,8,9,10. This cleavage results in the production of two mRNA fragments, a 5′-NGD intermediate and a 3′-NGD intermediate, which are further degraded by the exosome and Xrn1 exoribonuclease, respectively. The 5′-NGD intermediate lacks a termination codon and a ribosome translating a 5′-NGD intermediate may be stalled at the 3′ end. For a 5′-NGD intermediate to be rapidly degraded by the exosome, the stalled ribosome at the 3′ end of the mRNA fragment must be dissociated.
Dom34-Hbs1 has been proposed to be involved in the endonucleolytic cleavage of mRNA induced by blockage of translation elongation in vivo6,7,9,10,11. Recent biochemical analyses clearly demonstrated that yeast Dom34-Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off12,13. In yeast in vitro reconstituted translation system, Dom34-Hbs1 promotes the dissociation of the translation elongation complex into subunits, release of mRNA and drop-off of peptidyl-tRNA12,13. Mammalian Pelota-human Hbs1 (hHbs1) also induces the dissociation of the translation elongation complex, but only in the presence of ABCE1 and Hbs1 has only a stimulatory effect14. A recent study showed that the yeast Dom34-Hbs1 complex dissociates a ribosome stalled at the 3′ end of a 5′-NGD intermediate and stimulates the exosomal degradation of the 5′-NGD intermediate in vivo10. The Nonstop Decay (NSD) quality control system eliminates nonstop mRNA and Dom34-Hbs1 also facilitates the degradation of nonstop mRNAs from their 3′ ends by exosome10,15. These results indicate that the Dom34-Hbs1 complex plays a crucial role in both NGD and NSD by stimulating the degradation of this mRNA by dissociating the ribosome that is stalled at the 3′ end of nonstop mRNA. In dom34Δ mutant cells, a stalled ribosome at the 3′ end of nonstop mRNA may inhibit the degradation of nonstop mRNA by exosomes but induce sequential endonucleolytic cleavages. The mechanisms and physiological roles of sequential cleavages induced by stalled ribosome still remain elusive.
Nascent peptide-dependent translation arrest is crucial for quality control of eukaryotic gene expression. A genetic screen to identify factors required for induction of translation arrest by poly-arginine sequences revealed that RACK1 (ASC1 in yeast) is required for translation arrest9. Asc1/RACK1, which is highly conserved in eukaryotes, serves as a scaffold protein in many different signal transduction pathways. It also functions in developmental processes, such as sexual differentiation, cell-proliferation control, control of post-synaptic excitation in the brain and in hormone response pathways16,17,18,19. Asc1/RACK1 stably associates with the 40S subunit in a stoichiometric manner in yeast20,21,22. However, its role in translational control is still largely unknown. Two important questions to be resolved are how Asc1/RACK1 functions during translation arrest; how it can sense the properties of the nascent peptide in the ribosomal tunnel on the 60S subunit. Cryo-electron microscopy (Cryo-EM) structure of yeast ribosome revealed that the structure of the 60S subunit is affected by Asc1/RACK1 depletion23,24. Crystal structure studies showed that RACK1 binds to the head region of the 40S subunit in the vicinity of the mRNA exit channel21,22. These suggest that binding of Asc1/RACK1 to the 40S subunit stabilizes the conformation and/or the monitoring of the properties of nascent peptide in the ribosome tunnel by the 80S ribosome. It is also possible that recruitment of signaling factors through interactions with Asc1/RACK1 on the 40S subunit are required to modulate the activity of the ribosome or translation factors. A recent study showed that Asc1/RACK1 inhibits ribosomal +1 frameshifting at tandem CGA rare codons25. These findings suggest that Asc1/RACK1 may also modulate translation elongation at specific sites, implying that it plays an important and so far unappreciated role in the control of gene expression in eukaryotes.
Here, we describe the mechanism and biological relevance of a novel quality control system induced by stalled ribosomes at the 3′ end of mRNA. Asc1/RACK1 stimulated the degradation of truncated GFP-Rz mRNA that lacks both a termination codon and a poly(A) tail in dom34Δasc1Δ mutant cells. The sequential endonucleolytic cleavages by stalled ribosomes at the 3′ end of mRNA were drastically decreased by the deletion of ASC1/RACK1. Small fragments of truncated GFP-Rz mRNA were purified with stalled ribosomes in an Asc1/Rack1 dependent manner. Overproduction of nonstop mRNA inhibited the growth of the dom34Δasc1Δ mutant, possibly due to the elevated level of peptidyl-tRNA derived from truncated GFP-Rz mRNA. Moreover, the level of the peptidyl-tRNA was drastically increased by the deletion of Asc1/RACK1 in dom34Δ mutant background. Finally, we found that arrest products from truncated GFP-Rz mRNA in asc1Δ and dom34Δ mutant cells were degraded by Ltn1 that is an E3 ubiquitin ligase for co-translational degradation of arrest products. Based on our findings, we propose that Asc1/RACK1 stalls ribosome during aberrant translation elongation to induce endonucleolytic cleavages thereby reducing the level of aberrant mRNAs.
Results
Asc1/RACK1 facilitates the degradation of truncated GFP-Rz mRNA in the dom34Δ mutant background.
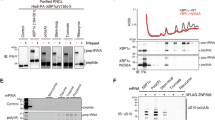
Previous work showed that Dom34-Hbs1 stimulates degradation of nonstop mRNA from the 3′ end10 (Fig. 1A). To determine whether Asc1/RACK1 is involved in NSD, we examined its role in the degradation of GFP-Rz mRNA in the absence of Dom34 (Fig. 1B). GFP-Rz mRNA is produced from GFP-Rz-FLAG-HIS3 mRNA by the self-cleavage activity of hammerhead ribozyme(Rz), therefore this mRNA lacks a poly(A) tail and a termination codon. As previously reported, GFP-Rz mRNA was significantly more stable when Dom34 and Hbs1 were absent from the xrn1Δ mutant, in which only the 3′-to-5′ degradation pathway is active10 (Fig. 1B, t1/2 = 4.9 min. for xrn1Δ vs. t1/2 = 27.6 min. for xrn1Δdom34Δ). We found that Asc1/RACK1 destabilized GFP-Rz mRNA in the absence of Dom34 (Fig. 1B, t1/2 = 4.1 min. for dom34Δ vs. t1/2 = 7.0 min. for dom34Δasc1Δ). Asc1/RACK1 also destabilized GFP-Rz mRNA in ski2Δdom34Δ mutant background (Fig. 1B, t1/2 = 5.0 min. for ski2Δdom34Δ vs. t1/2 > 32 min. for asc1Δski2Δdom34Δ). These indicate that Asc1/RACK1 facilitates truncated GFP-Rz mRNA decay in the absence of exosome-dependent decay and Dom34.
Asc1/RACK1 facilitates truncated GFP-Rz mRNA decay in the absence of 5′- to -3′ decay pathway.
(A) Quality control of ribosome stalled at the 3′ end of truncated GFP-Rz mRNA. In the ribosome-associated quality control (RQC) system, a stalled ribosome dissociates into subunits by the functions of Dom34-Hbs1 in concert with Rli1 and the peptidyl-tRNA on the 60S subunit is ubiquitylated by Ltn1 Listerin in mammal to target it for proteasomal degradation. In NSD, Dom34-Hbs1 dissociates the stalled ribosome from nonstop mRNA, thereby facilitating degradation of nonstop mRNA by the exosome. (B) Asc1/RACK1 facilitates degradation of GFP-Rz mRNA in the absence of Dom34. (Top) Schematic drawing of truncated GFP-Rz mRNA derived from the GFP-Rz-FLAG-HIS3 reporter gene. GFP-Rz mRNA, which lacks a poly(A) tail and a termination codon, is produced from GFP-Rz-FLAG-HIS3 mRNA by the self-cleavage activity of hammerhead ribozyme (Rz). DIG-labeled G3 probe corresponding to 654–714 nt of GFP is indicated. (Bottom) The stability of GFP-Rz mRNA in the indicated mutant cells. Cells were grown in SC-Ura media containing 2%w/v galactose and transcription was inhibited by addition of 2%w/v glucose at mid-log phase. At the indicated times, cells were harvested and the relative levels of truncated GFP-Rz mRNA were determined by Northern blot analysis using a DIG-labeled G3 probe.
Asc1/RACK1 is required for endonucleolytic cleavage of truncated GFP-Rz mRNA in the dom34Δ mutants.
We previously reported that Asc1/RACK1 is involved in NGD, endonucleolytic cleavage induced by translation arrest9. Therefore, we hypothesized that sequential endonucleolytic cleavages destabilize GFP-Rz mRNA in ski2Δdom34Δ mutant cells and Asc1/RACK1 is required for the sequential endonucleolytic cleavages induced by the stalled ribosome at the 3′ end of GFP-Rz mRNA in ski2Δdom34Δ mutant cells. To test the possibility that Asc1/RACK1 is required for the sequential endonucleolytic cleavages induced by the stalled ribosome, we determined the levels of 40–200 nt fragments derived from GFP-Rz reporter mRNAs by Northern blot analysis, as previously described10 (Fig. 2A). Upon deletion of ASC1/RACK1, the levels of fragments derived from GFP-Rz mRNA were drastically reduced in dom34Δmutant cells (Fig. 2B, lanes 8 and 9), ski2Δdom34Δ mutant cells (Fig. 2B, lanes 10 and 11) or xrn1Δdom34Δ mutant cells (Fig. 2B, lanes 12 and 13). Deletion of ASC1/RACK1 diminished the production of fragments in ski2Δdom34Δ and xrn1Δdom34Δ mutant cells (Fig. 2C). These findings indicate that Asc1/RACK1 is generally required for the sequential endonucleolytic cleavages induced by stalled ribosomes at the 3′ ends of mRNA.
Asc1/RACK1 is required for sequential endonucleolytic cleavages induced by stalled ribosomes.
(A) Multiple endonucleolytic cleavages of truncated GFP-Rz mRNA in the dom34Δ mutant background. (B) Asc1/RACK1 is required for multiple endonucleolytic cleavages of GFP-Rz mRNA induced by stalled ribosomes. RNA samples were prepared from the indicated cells harboring p416GAL1p-GFP-Rz-FLAG-HIS3 and analyzed by 1.2% agarose and Northern blotting using DIG-labeled G3 and SCR1 probes. The levels of GFP-Rz mRNA and fragments were determined using the samples from ski2Δdom34Δasc1Δ cells harboring p416GPDp-GFP-Rz-FLAG-HIS3 as a standard curve (lanes 1–5). The relative levels of each mRNA or fragments were normalized with respect to the GFP-Rz mRNA level in wild-type cells, which was assigned a value of 100 and reported as the mean values from three independent experiments. (C) Asc1/RACK1 is required for multiple endonucleolytic cleavages of GFP-Rz mRNA induced by stalled ribosomes. RNA samples were prepared from the indicated cells harboring p416GAL1p-GFP-Rz-FLAG-HIS3 and analyzed by 6% TBE-Urea PAGE and Northern blotting using DIG-labeled G3 and SCR1 probes.
Small fragments of truncated GFP-Rz mRNA were purified with stalled ribosomes in an Asc1/Rack1 dependent manner.
To confirm that stalled ribosome at the 3′ end of mRNA in dom34Δ mutant cells contains small RNA fragments, we purified stalled ribosomes from cells expressing both Protein A-TEV-GFP-Rz and Rps2-FLAG proteins as depicted in Fig. 3A. The Protein A-TEV-GFP-Rz truncated products derived from GFP-Rz truncated mRNA were purified with IgG affinity beads and eluted by the addition of TEV protease. Then stalled ribosomes were affinity-purified with anti-FLAG resin. The stalled ribosomes containing peptidyl-tRNA were purified from both dom34Δ mutant and asc1Δdom34Δ mutant cells with this (Fig. 3B,C). Northern blot analysis of the purified samples with GFP G3 probe clearly revealed that the purified stalled ribosomes from dom34Δ cells contain fragments derived from GFP-Rz mRNA (Fig. 3D, lane 3). In contrast, stalled ribosomes from asc1Δdom34Δ cells contain less amounts of small fragments derived from GFP-Rz mRNA and the levels of fragments were the same as that from dom34Δ mutant cells (Fig. 3D). These results clearly indicate that stalled ribosomes at the 3′ end of GFP-Rz mRNA in dom34Δ mutant cells contain fragments produced by sequential endonucleolytic cleavages in an Asc1/RACK1-dependent manner.
Purification of stalled ribosome at the 3′ end of GFP-Rz truncated mRNA.
(A) Schematic illustration of the split-tag affinity purification of the stalled ribosome. (B) Cells harboring pGPDp-ProteinA-TEV-GFP-Rz-HIS3 were harvested and cell lysates prepared for split-tag purification. Protein samples of the purified fractions were analyzed by CBB stain (lanes 1–2). Ribosomes were affinity-purified from Rps2-FLAG cells as a control (lane 3). (C) Protein samples of the purified fractions were analyzed by NuPAGE followed by Western blot analysis with anti-GFP antibody (Top panel) or anti-FLAG, anti-RpL23 and anti-Asc1 antibodies (Bottom panles). (D) RNA samples of the purified fractions were analyzed by Northern blotting using DIG-labeled G3 probes after PAGE.
An endonucleolytic cleavage induced by stalled ribosome requires ribosome-associated Asc1/RACK1
We previously reported that Asc1/RACK1 participates in nascent peptide-dependent translation arrest and that the binding of Asc1/RACK1 to the 40S subunit is crucial for this translation arrest. The asc1D109Y and asc1R38DK40E mutants were found to produce severe defects both in translation repression by polybasic sequence and in the association between RACK1 and the ribosome9. We introduced mutations in residues located at the sites of interaction of Asc1/RACK1 with the 40S subunit26 (Fig. 4A, pink residues). We determined the levels of the full-length products derived from GFP-R12-FLAG-HIS3 (R12) reporter and three Asc1/RACK1 mutant proteins (16HNG18AAA, 38RDK40AAA and 85WDK87AAA) were defective in translation arrest induced by a polybasic amino-acid sequence (Fig. 4B). Western blot analysis followed by centrifugation in sucrose density gradients revealed that these three Asc1/RACK1 mutant proteins (16HNG18AAA, 38RDK40AAA and 85WDK87AAA) were defective in interaction with ribosome (Fig. 4C). These results are consistent with previous results showing that the binding of Asc1/RACK1 to the 40S subunit is crucial for translation arrest induced by polybasic sequence9. We next examined whether the 40S subunit binding of Asc1/RACK1 is crucial for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Production of fragments was diminished in ski2Δdom34Δ and xrn1Δdom34Δ mutant cells expressing these Asc1/RACK1 mutants (Fig. 4D). These indicate that the Asc1/RACK1-bound ribosomes are competent to induce the sequential endonucleolytic cleavages induced by stalled ribosomes at the 3′ end of mRNA.
Ribosome binding activity of Asc1/RACK1 is required for sequential endonucleolytic cleavages induced by stalled ribosomes.
(A) The structure diagram was prepared in PyMOL using PDB file 4V88. (Left) Residues located in the putative interaction sites of Asc1/RACK1 with the 40S subunit are shown in pink. Residues replaced with alanine residues in mutants defective in translation arrest by polybasic sequence and the interaction with 40S subunit were shown in Red. (Middle) The residues involved in translation arrest by polybasic sequence and the interaction with 40S subunit locate close to H39-H40 and Rps17. (Right) Relative position of D109 of Asc1 to R33 of Rps17. (B) The ribosome binding of Asc1/RACK1 mutant proteins defective in translation repression by polybasic amino-acid sequence. The asc1Δ cells harboring the GFP-R12-FLAG-HIS3 reporter were transformed with the various FLAG-Asc1/RACK1 plasmids. The levels of the GFP-R12-FLAG-HIS3 reporter proteins were determined by Western blot analysis with anti-GFP and anti-FLAG antibodies. (C) Ribosome binding of Asc1/RACK1 mutant proteins. Cell extracts were prepared from the asc1Δ cells expressing various FLAG-Asc1/RACK1 mutant proteins. Extracts were subjected to polysome analysis, followed by Western blot analysis with anti-FLAG antibody. (D) Ribosome-associated Asc1/RACK1 is required for multiple endonucleolytic cleavages of GFP-Rz mRNA in a dom34Δ mutant background. ski2Δdom34Δasc1Δ mutant cells harboring p416GAL1p-GFP-Rz-FLAG-HIS3 were transformed with the indicated plasmids expressing Asc1/RACK1 mutants. RNA samples were analyzed by 6% TBE-Urea PAGE and Northern blotting using DIG-labeled G3 probes.
Overproduction of truncated GFP-Rz mRNAs inhibits cell growth of the ski2Δdom34Δasc1Δ mutant.
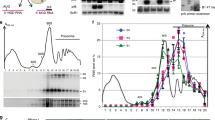
Dom34-Hbs1 promotes dissociation of the translation elongation complex into subunits and we detected peptidyl-tRNA derived from GFP-Rz mRNA in cell extracts of the ski2Δdom34Δ mutant10. Although GFP-Rz mRNAs were mainly distributed in heavy polysome fractions, the peptidyl-tRNA is only present in monosome or light polysome fractions10. Because the sequential endonucleolytic cleavages were almost abolished in the absence of ASC1/RACK1, we had expected to detect the peptidyl-tRNA derived from GFP-Rz mRNA in heavy polysome fractions derived from cell extracts of the ski2Δdom34Δasc1Δ mutant. However, in the ski2Δdom34Δasc1Δ mutant, the peptidyl-tRNA derived from GFP-Rz mRNA was distributed in the monosome and light polysome fractions and the distribution of peptidyl-tRNA was almost the same as that in ski2Δdom34Δ mutant cells (Fig. 5A). The levels of peptidyl-tRNA in polysome fractions and the polypeptide in ribosome-free fractions were significantly increased by ASC1/RACK1 deletion in the ski2Δdom34Δ mutant background (Fig. 5A). Therefore, we quantified the levels of peptidyl-tRNA and free polypeptide derived from GFP-Rz mRNA in cell lysates (Fig. 5B). Neutral-PAGE (NuPAGE) followed by Western blot analysis revealed that the levels of peptidyl-tRNA and free polypeptide derived from GFP-Rz mRNA were drastically increased by ASC1/RACK1 deletion in the ski2Δdom34Δ mutant background. The level of peptidyl-tRNA in ski2Δdom34Δasc1Δ mutant was 9-fold higher than that in ski2Δdom34Δ mutant cells and the level of free polypeptide GFP-Rz in the ski2Δdom34Δasc1Δ mutant was 24-fold higher than that in ski2Δdom34Δ mutant cells (Fig. 5B). These findings indicate that nonstop protein may be synthesized from GFP-Rz mRNA even in the absence of the Dom34-Hbs1 complex. Therefore, we propose that ribosomes stalled at the 3′ end of mRNA in the absence of Dom34-Hbs1 may be dissociated into subunits in the asc1Δ mutant background. To determine the biological significance of this novel quality control system, we examined the effects of overproduction of aberrant mRNAs without termination codons in both ski2Δdom34Δasc1Δ mutant and ski2Δdom34Δ mutant. To overproduce GFP-Rz mRNA, cells harboring pGAL1p-GFP-Rz-FLAG-HIS3 plasmid were grown on SC-Ura plates containing galactose. Overproduction of GFP-Rz nonstop mRNA inhibited growth of the ski2Δdom34Δasc1Δ mutant, but not the ski2Δdom34Δ mutant (Fig. 5C). We propose that when the quality control system by Dom34-Hbs1 is impaired, Asc1/RACK1 plays a crucial role in the repression of aberrant products derived from mRNAs lacking termination codons by inducing sequential endonucleolytic cleavages.
Asc1/RACK1 inhibits protein production from nonstop mRNA in the absence of Dom34.
(A) Cell extracts were prepared from the indicated cells harboring p416GPDp-GFP-Rz-FLAG-HIS3 plasmid and polysome analysis was performed. (Top) Absorbance at 254 nm. (Middle) Neutral-PAGE followed by Western blotting with anti-GFP antibodies. (Bottom) Northern blotting using DIG-labeled G3 probes. (B) Levels of peptidyl-tRNA derived from GFP-Rz mRNA in ski2Δdom34Δ or ski2Δdom34Δasc1Δ mutant cells. Cell extracts were analyzed by Neutral-PAGE, followed by Western blotting using anti-GFP and anti-eEF2 antibodies. The levels of GFP-Rz and peptidyl-tRNA were determined using the samples from ski2Δdom34Δasc1Δ cells harboring p416GPDp-GFP-Rz-FLAG-HIS3 as a standard curve (lanes 1–6). The relative levels of each GFP-Rz and peptidyl-tRNAs were normalized with respect to the levels of GFP-Rz and peptidyl-tRNAs in wild-type cells, which was assigned as a value of 100 and reported as the mean values from three independent experiments. (C) Overproduction of aberrant mRNAs lacking a termination codon inhibits growth of the ski2Δdom34Δasc1Δ mutant. Cells harboring pGAL1p-GFP-Rz-FLAG-HIS3 were grown in SC-Ura media containing 2% raffinose to log-phase and the cells in a series of dilutions were grown on SC-Ura plates containing 2% glucose or 2% galactose.
Arrest products from truncated GFP-Rz mRNA were subjected to Ltn1-dependent degradation in asc1 and dom34 mutant cells.
Co-translational degradation of polypeptide nascent chains plays a critical role in quality control of protein synthesis and the rescue of stalled ribosomes. Nonstop proteins are recognized by the ribosome-associated quality control (RQC) machinery and targeted for proteasomal degradation27,28,29,30,31,32. Dom34-Hbs1 and its mammalian homolog Pelota-hHbs1 is involved in RQC by facilitating the dissociation of stalled ribosomes at the 3′ end of mRNA32. We previously reported that the level of the protein products derived from the reporter gene containing poly-lysine sequences within the ORF was drastically increased in asc1Δ mutant, whereas the levels of arrest products were significantly decreased in asc1Δ mutant even in the presence of the proteasome inhibitor9. Therefore, we determined the levels of GFP-Rz and the peptidyl-tRNA in dom34Δ and dom34Δasc1Δ mutant cells. Ltn1 significantly decreased the levels of Nonstop products in dom34Δmutant cells (Fig. 6A, lanes 1 and 3) and dom34Δasc1Δ mutant cells (Fig. 6A, lanes 2 and 4). The ATPase activity of the Ski2-3-8 complex was abolished by E445Q mutation in the Ski2 DExH core33. The level of GFP-Rz mRNA was increased in the presence of Ski2-E445Q mutant protein in wild type cells (Fig. 6B). Because of the problems during the construction of ski2Δasc1Δltn1Δdom34Δ tetra mutants, we introduced the dominant negative Ski2-E445Q mutant to inhibit the exosome activity33. In the presence of Ski2-E445Q mutant protein, Ltn1 also decreased the levels of GFP-Rz and the peptidyl-tRNA in dom34Δ and dom34Δasc1Δ mutant cells (Fig. 6A, lanes 5–8). These indicate that Ltn1 degrades the arrest products from truncated GFP-Rz mRNA even in the absence of Asc1 and Dom34.
Nonstop products derived from truncated GFP-Rz mRNA were degraded by RQC in dom34 and dom34asc1 mutant cells.
(A) Levels of peptidyl-tRNA and GFP-Rz protein derived from GFP-Rz mRNA were significantly increased by the deletion of Ltn1 in dom34Δ or dom34Δasc1Δ mutant cells. Cell extracts prepared from the indicated cells containing GFP-Rz-FLAG-HIS3 reporter were analyzed by Neutral-PAGE, followed by Western blotting using anti-GFP antibody. When indicated, cells were transformed with Ski2 E445Q plasmid (Lanes 5–8). (B) The level of truncated GFP-Rz mRNA was increased in wild type cells in the presence of dominant-negative Ski2-E445Q protein. (C) A model for quality control of ribosomes stalled at 3′ ends of nonstop mRNAs in dom34Δ and dom34Δasc1Δ mutant. (Top) In the absence of Dom34, a stalled ribosome at the 3′ end of a nonstop mRNA induces sequential cleavage, resulting in production of a ladder of small fragments protected by ribosomes. (Bottom) In the absence of Dom34 and Asc1/RACK1, a ribosome reaching the 3′ end of a nonstop mRNA dissociates from mRNA as an 80S particle containing peptidyl-tRNA. In both mutant cells, Ltn1 degrades nonstop products derived from GFP-Rz mRNA.
Discussion
The Dom34-Hbs1 complex dissociates ribosomes stalled at the 3′ ends of 5′-NGD intermediates and nonstop mRNAs and stimulates their degradation by the exosome10. In dom34Δ mutant cells, such stalled ribosomes induce sequential endonucleolytic cleavages10. These cleavages have been proposed to play novel roles in NGD and NSD by reducing the levels of 5′-NGD intermediates or nonstop mRNA in the absence of Dom34-Hbs1-dependent mRNA decay mediated by the exosome10. The results in this study clearly showed that Asc1/RACK1 is required for sequential endonucleolytic cleavages induced by stalled ribosome at the 3′ end of truncated GFP-Rz mRNA. Asc1/RACK1 also was reported to facilitate the endonucleolytic cleavages induced by translation arrests by polybasic sequences9. Therefore we propose that Asc1/RACK1 is a quality control factor to stimulate the endonucleolytic cleavages by stalled ribosome generally.
The level of GFP-Rz mRNA in ski2Δdom34Δasc1Δ mutant was 2.5-fold higher than that in ski2Δdom34Δmutant cells (Fig. 2B). The levels of peptidyl-tRNA and GFP-Rz derived from truncated GFP-Rz mRNA were increased more than 9-fold and 24-fold respectively, by ASC1/RACK1 deletion in the ski2Δdom34Δ mutant background (Fig. 5B). Therefore, translation efficiency based on the ratio between protein products and mRNA levels was significantly increased by ASC1/RACK1 deletion in the ski2Δdom34Δ mutant background. Moreover, Asc1/RACK1 moderately but significantly stabilized GFP-Rz mRNA in xrn1Δdom34Δ mutant background (Fig. 1B, t1/2 = 27.6 min. for xrn1Δdom34Δ vs. t1/2 = 18.2 min. for asc1Δxrn1Δdom34Δ), suggesting that Asc1/RACK1 facilitates the exosome-dependent decay from 3′ end of GFP-Rz mRNA. We suspect that ribosomes reaching the 3′ end of a nonstop mRNA rapidly dissociate as 80S particles containing peptidyl-tRNA in the absence of Dom34 and Asc1/RACK1, resulting in elevated levels of peptidyl-tRNA derived from nonstop mRNA. We previously reported that Asc1/RACK1 is required for translation arrest induced by polybasic amino-acid sequence or tandem rare codons9,34. Taken together, these observations suggest that Asc1/RACK1 plays general roles in translation regulation and mRNA quality control systems and that Asc1/RACK1-bound ribosomes have the potential to stall in response to specific nascent peptide sequences, low levels of aminoacyl-tRNA, or a lack of a codon at the A-site.
Recent study showed that Asc1 promotes translation of mRNAs with short open reading frames35. The D109Y mutant showed translational defects that, although correlated with those observed in the asc1Δ mutants were much smaller in magnitude. The R38DK40E mutant showed almost negligible effects on translation35. In contrast, the defects of Asc1 mutants (38RDK40AAA, 85WDK87AAA, D109Y) in translation arrest by polybasic sequence and the endonucleolytic cleavage induced by stalled ribosome were almost the same as that of the asc1Δ mutant, although 16HNG18AAA mutant showed moderate effects (Fig. 4). We propose that robust and proper interaction between Asc1/RACK1 with 40S subunit may be crucial for co-translational quality controls to induce translation elongation or mRNA cleavage, but dispensable to promote translation of mRNAs with short open reading frames. Drosophila RACK1 is required in the 40S ribosomal subunit for IRES-dependent translation of Cricket Paralysis Virus and the picorna-like Drosophila C Virus36. D108Y and R38D/K40E mutants were defective in these virus RNAs36, suggesting the regulatory roles of the interaction of Asc1/RACK1 with 40S subunit. The identification of the interacting partner for Asc1/RACK1 is crucial to elucidate mechanisms of RACK1-dependent regulation of translation or mRNA decay.
The results in this study clearly showed that Ltn1 destabilizes aberrant nonstop polypeptides even in the absence of the ribosome dissociation factor Dom34:Hbs1 as previously reported34. We also found that Ltn1 destabilizes nonstop products derived from GFP-Rz mRNA in the absence of ASC1/RACK1. These indicate that ASC1/RACK1 is dispensable for RQC for nonstop products. Previous results showed that Ltn1 destabilizes arrest products derived from GFP-R12-FLAG-HIS3 reporter in a RACK1-dependent manner34, indicating that the roles of Asc1/RACK1 in RQC differs depending on where ribosomes stalled. Mechanisms of the dissociation of stalled ribosomes are still largely unknown and future study will identify putative factors that recognize stalled ribosome and induce the subunit dissociation independent of Dom34-Hbs1.
Methods
Strains and other methods
The strains and plasmids used in this study are listed in Supplementary Table 1. Northern blot analysis, protein preparation, Western blot analysis were performed as previously described10,27.
Yeast extract and sucrose gradient separation
Yeast cells were grown exponentially at 30 °C and harvested by centrifugation. Cell extracts were prepared as described previously20. The equivalent of 50 A260 units were layered onto linear 10–50% sucrose density gradients. Sucrose gradients (10–50% sucrose in 10 mM Tris-acetate pH 7.4, 70 mM ammonium acetate and 4 mM magnesium acetate) were prepared in polyallomer tubes (Beckman Coulter) using a Gradient Master. Crude extracts were layered on top of the sucrose gradients and centrifuged at 150,000 × g in a P28S rotor (Hitachi Koki, Japan) for 3 hours at 4 °C. Gradients were then fractionated (TOWA lab, Tsukuba). Polysome profiles were generated by continuous absorbance measurement at 254 nm using a single path UV-1 optical unit (ATTO BioMini UV-monitor) connected to a chart recorder (ATTO digital mini-recorder). Equal volume fractions were collected and processed for Northern or Western blotting as described above37.
Determination of mRNA stability
Yeast cells were grown in minimal medium (SC medium) containing 2% galactose. Cells were grown to OD600 = 0.6, harvested and resuspended in medium containing 2% glucose to inhibit transcription from the GAL1 promoter. At the indicatedtimes, the cells were harvested to prepare RNA samples by acidic phenol RNA extraction method using water-saturated phenol and phenol-chloroform-isoamylalcohol (25:24:1) mixture. mRNA levels of reporter genes were determined by Northern blotting using digoxigenin (DIG)-labeled GFP probes prepared by PCR-based nucleic acid labeling using PCR DIG Probe Synthesis kit (Roche, NJ, USA) according to the procedure specified by the manufacturer and primers 5′-GCTCTAGAATGAGTAAAGGAGAAGAACTTTTCAC-3′ and 5′-GGACTAGTTTTGTATAGTTCATCCATGCCA-3′. GFP G3 probe is a 5′ end DIG-labeled oligonucleotide 5′-TTTGTATAGTTCATCCATGCCATGTGTAATCCCAGCAGCAGTTACAAACTCAAGAAGGACC-3′. The intensity of bands on the blots was quantified on a LAS4000 mini (GE Healthcare). Relative RNA levels were determined using Multi Gauge v3.0 (Fujifilm, Japan) by comparison to a standard curve using a series of dilutions of samples from time 0 (just before the addition of glucose).
Split-tag affinity purification of stalled ribosomes
Stalled ribosomes were purified from cells expressing both ProtA-TEV-GFP-Rz-His3 containing and Rps2-FLAG proteins by split-tag affinity purification. Yeast cells were transformed with plasmids harbouring ProteinA-TEV-GFP-Rz-HIS3 gene were cultured in 1 L of synthetic complete medium. The ProteinA-TEV-GFP-Rz was affinity-purified from whole cell lysates using IgG SepharoseTM (GE Healthcare), followed by the treatment with TEV protease. Ribosome complex containing ProteinA-TEV-GFP-Rz was purified with Anti-DYKDDDDK tag antibody beads (Wako, Japan) and eluted by 250ng/μL of FLAG peptides.
Detection of peptidyl-tRNA
Cell extracts or polysome fractions were analyzed by NuPAGE (Invitrogen, USA) followed by Western blotting with anti-GFP antibodies10. To destroy the RNA moiety of peptidyl-tRNA, RNase A was added to the samples at a final concentration of 10 mg/l and the samples were incubated at 37 °C for 10 min. (data not shown).
Spot assay
Cells harboring p416GAL1p-GFP-Rz-FLAG-HIS3 plasmids were grown with SC-Ura media containing 2% raffinose to OD600 = 0.3. The cells were spotted on SC-Ura plate containing 2% glucose or 2% galactose after a series of 10-fold dilution and incubated at 30 °C for 2 days (glucose) or 3 days (galactose).
Overexpression of dominant-negative Ski2 and inhibition of exosome
The ATPase activity of the Ski2-3-8 complex was abolished by E445Q mutation in the Ski2 DExH core33. To inhibit exosome activity, p415GPDp-SKI2 E445Q plasmids were introduced to cells. Inhibition of exosome was confirmed by northern blot to detect GFP-Rz mRNA.
Additional Information
How to cite this article: Ikeuchi, K. and Inada, T. Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Sci. Rep. 6, 28234; doi: 10.1038/srep28234 (2016).
References
Schoenberg, D. R. & Maquat, L. E. Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13, 246–259, doi: 10.1038/nrg3160 (2012).
Pechmann, S., Willmund, F. & Frydman, J. The ribosome as a hub for protein quality control. Mol Cell 49, 411–421, doi: 10.1016/j.molcel.2013.01.020 (2013).
Shao, S. & Hegde, R. S. Target Selection during Protein Quality Control. Trends Biochem Sci 41, 124–137, doi: 10.1016/j.tibs.2015.10.007 (2016).
Brandman, O. & Hegde, R. S. Ribosome-associated protein quality control. Nat Struct Mol Biol 23, 7–15, doi: 10.1038/nsmb.3147 (2016).
Lykke-Andersen, J. & Bennett, E. J. Protecting the proteome: Eukaryotic cotranslational quality control pathways. J Cell Biol 204, 467–476, doi: 10.1083/jcb.201311103 (2014).
Doma, M. K. & Parker, R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564, doi: 10.1038/nature04530 (2006).
Passos, D. O. et al. Analysis of Dom34 and its function in no-go decay. Mol Biol Cell 20, 3025–3032, doi: 10.1091/mbc.E09-01-0028 (2009).
Harigaya, Y. & Parker, R. No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip Rev RNA 1, 132–141, doi: 10.1002/wrna.17 (2010).
Kuroha, K. et al. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep 11, 956–961, doi: 10.1038/embor.2010.169 (2010).
Tsuboi, T. et al. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell 46, 518–529, doi: 10.1016/j.molcel.2012.03.013 (2012).
van den Elzen, A. M. et al. Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nat Struct Mol Biol 17, 1446–1452, doi: 10.1038/nsmb.1963 (2010).
Shoemaker, C. J., Eyler, D. E. & Green, R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330, 369–372, doi: 10.1126/science.1192430 (2010).
Shoemaker, C. J. & Green, R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA 108, E1392–1398, doi: 10.1073/pnas.1113956108 (2011).
Pisareva, V. P., Skabkin, M. A., Hellen, C. U., Pestova, T. V. & Pisarev, A. V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 30, 1804–1817, doi: 10.1038/emboj.2011.93 (2011).
Saito, S., Hosoda, N. & Hoshino, S. I. Hbs1-Dom34 functions in non-stop mRNA decay (NSD) in mammalian cells. J Biol Chem, doi: 10.1074/jbc.M112.448977 (2013).
Guo, J. et al. Dissection of the relationship between RACK1 and heterotrimeric G-proteins in Arabidopsis. Plant Cell Physiol 50, 1681–1694, doi: 10.1093/pcp/pcp113 (2009).
Volta, V. et al. RACK1 depletion in a mouse model causes lethality, pigmentation deficits and reduction in protein synthesis efficiency. Cell Mol Life Sci 70, 1439–1450, doi: 10.1007/s00018-012-1215-y (2013).
Yaka, R. et al. NMDA receptor function is regulated by the inhibitory scaffolding protein, RACK1. Proc Natl Acad Sci USA 99, 5710–5715, doi: 10.1073/pnas.062046299 (2002).
Kadrmas, J. L., Smith, M. A., Pronovost, S. M. & Beckerle, M. C. Characterization of RACK1 function in Drosophila development. Dev Dyn 236, 2207–2215, doi: 10.1002/dvdy.21217 (2007).
Inada, T. et al. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA 8, 948–958 (2002).
Coyle, S. M., Gilbert, W. V. & Doudna, J. A. Direct link between RACK1 function and localization at the ribosome in vivo. Mol Cell Biol 29, 1626–1634, doi: 10.1128/MCB.01718-08 (2009).
Rabl, J., Leibundgut, M., Ataide, S. F., Haag, A. & Ban, N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331, 730–736, doi: 10.1126/science.1198308 (2011).
Nilsson, J., Sengupta, J., Frank, J. & Nissen, P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep 5, 1137–1141, doi: 10.1038/sj.embor.7400291 (2004).
Sengupta, J. et al. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol 11, 957–962, doi: 10.1038/nsmb822 (2004).
Wolf, A. S. & Grayhack, E. J. Asc1, homolog of human RACK1, prevents frameshifting in yeast by ribosomes stalled at CGA codon repeats. RNA 21, 935–945, doi: 10.1261/rna.049080.114 (2015).
Taylor, D. J. et al. Comprehensive molecular structure of the eukaryotic ribosome. Structure 17, 1591–1604, doi: 10.1016/j.str.2009.09.015 (2009).
Ito-Harashima, S., Kuroha, K., Tatematsu, T. & Inada, T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev 21, 519–524, doi: 10.1101/gad.1490207 (2007).
Dimitrova, L. N., Kuroha, K., Tatematsu, T. & Inada, T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem 284, 10343–10352, doi: 10.1074/jbc.M808840200 (2009).
Brandman, O. et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054, doi: 10.1016/j.cell.2012.10.044 (2012).
Shen, P. S. et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78, doi: 10.1126/science.1259724 (2015).
Shao, S., Brown, A., Santhanam, B. & Hegde, R. S. Structure and assembly pathway of the ribosome quality control complex. Mol Cell 57, 433–444, doi: 10.1016/j.molcel.2014.12.015 (2015).
Shao, S. & Hegde, R. S. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol Cell 55, 880–890, doi: 10.1016/j.molcel.2014.07.006 (2014).
Halbach, F., Reichelt, P., Rode, M. & Conti, E. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell 154, 814–826, doi: 10.1016/j.cell.2013.07.017 (2013).
Matsuda, R., Ikeuchi, K., Nomura, S. & Inada, T. Protein quality control systems associated with no-go and nonstop mRNA surveillance in yeast. Genes Cells 19, 1–12, doi: 10.1111/gtc.12106 (2014).
Thompson, M. K., Rojas-Duran, M. F., Gangaramani, P. & Gilbert, W. V. The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. Elife 5, doi: 10.7554/eLife.11154 (2016).
Majzoub, K. et al. RACK1 controls IRES-mediated translation of viruses. Cell 159, 1086–1095, doi: 10.1016/j.cell.2014.10.041 (2014).
Inada, T. & Aiba, H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J 24, 1584–1595, doi: 10.1038/sj.emboj.7600636 (2005).
Acknowledgements
We thank members of our laboratories for discussion and critical comments on the manuscript. K.I. was a recipient of a JSPS Research Fellowship. This study was supported by a Grant-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science (grant numbers 22370062 and 26116003) and by Research Grants in the Natural Sciences from the Takeda Foundation (to TI).
Author information
Authors and Affiliations
Contributions
K.I. performed experiments and data analysis. T.I. designed the study, performed data analysis. K.I. and T.I. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Ikeuchi, K., Inada, T. Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Sci Rep 6, 28234 (2016). https://doi.org/10.1038/srep28234
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep28234
This article is cited by
-
Molecular interactions between Hel2 and RNA supporting ribosome-associated quality control
Nature Communications (2019)
-
Ribothrypsis, a novel process of canonical mRNA decay, mediates ribosome-phased mRNA endonucleolysis
Nature Structural & Molecular Biology (2018)
-
Modulating cellular balance of Rps3 mono-ubiquitination by both Hel2 E3 ligase and Ubp3 deubiquitinase regulates protein quality control
Experimental & Molecular Medicine (2017)
-
Preribosomes escaping from the nucleus are caught during translation by cytoplasmic quality control
Nature Structural & Molecular Biology (2017)
-
Ubiquitination of stalled ribosome triggers ribosome-associated quality control
Nature Communications (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.