Abstract
The CRISPR/Cas9 system is an efficient and convenient tool for genome editing in plants. Cas9 nuclease derived from Streptococcus pyogenes (Sp) is commonly used in this system. Recently, Staphylococcus aureus Cas9 (SaCas9)-mediated genome editing was reported in human cells and Arabidopsis. Because SaCas9 (1053 a.a.) is smaller than SpCas9 (1368 a.a.), SaCas9 could have substantial advantages for delivering and expressing Cas9 protein, especially using virus vectors. Since the protospacer adjacent motif (PAM) sequence of SaCas9 (5′-NNGRRT-3′) differs from that of SpCas9 (5′-NGG-3′), the use of this alternative Cas9 nuclease could expand the selectivity at potential cleavage target sites of the CRISPR/Cas9 system. Here we show that SaCas9 can mutagenize target sequences in tobacco and rice with efficiencies similar to those of SpCas9. We also analyzed the base preference for ‘T’ at the 6th position of the SaCas9 PAM. Targeted mutagenesis efficiencies in target sequences with non-canonical PAMs (5′-NNGRRV-3′) were much lower than those with a canonical PAM (5′-NNGRRT-3′). The length of target sequence recognized by SaCas9 is one or two nucleotides longer than that recognized by SpCas9. Taken together, our results demonstrate that SaCas9 has higher sequence recognition capacity than SpCas9 and is useful for reducing off-target mutations in crop.
Similar content being viewed by others
Introduction
The clustered regularly interspaced short palindromic repeat (CRISPR)-associated endonuclease (CRISPR/Cas) system, the feature widely observed in prokaryote genomes, functions as an adaptive immune system by installing short DNA fragments from viruses or plasmids into CRISPR loci using RNA from the CRISPR loci to target specific sequences in invader genomes1,2. Of the three types of CRISPR/Cas systems, type II has been most intensively studied and developed for genome engineering techniques in eukaryotes, including plants3,4,5,6,7,8,9.
The type II CRISPR/Cas system is composed of a Cas9 nuclease and a guide RNA (gRNA)10,11. The chimeric single gRNA comprises two parts: a scaffold and a specific DNA target sequence on the 5′ end. The Cas9 nuclease most commonly used in genome editing research to date is derived from Streptococcus pyogenes (Sp)12. SpCas9 mediates genome editing at a site complementary to a 20-nucleotide DNA specific sequence in the gRNA13. Cas9 requires a protospacer adjacent motif (PAM) at the 3′ end of the DNA target sequence to recognize and cleave the target DNA14. The PAM sequence for SpCas9 is 5′-NGG-3′15.
Several Cas9 protein homologs have been identified and characterized for use in genome editing research16. Each Cas9 protein homolog functions similarly as an RNA-guided endonuclease, with homologs differing in their molecular weights and PAM preferences. Recently, Ran et al. reported that, when different Cas9 homologues were tested in mammalian cells, Staphylococcus aureus Cas9 (SaCas9) exhibited robust activity17. SaCas9 (1053 amino acids) is smaller than SpCas9 (1368 amino acids). The relatively large size of SpCas9 has limited its utility, thus the smaller Cas9 from Staphylococcus aureus might prove useful for therapeutic applications using the versatile adeno-associated virus delivery vehicle17. Unlike SpCas9, SaCas9 requires a 21- or 22-nt DNA specific sequence and the motif 5′-NNGRRT-3′ as a PAM sequence17. To broaden the spectrum of genome editing using the CRISPR/Cas9 system in plants, we need to widen the choice of Cas9 proteins beyond SpCas9.
During the preparation of this paper, Steinert et al. reported that SaCas9 could induce mutation in Arabidopsis thaliana18. Here, we show that SaCas9 functions in rice and tobacco. The probability of off-target mutation induced by SaCas9 might be lower than that induced by SpCas9. SaCas9 prefers the canonical PAM for effective targeted mutagenesis. Our results suggest that SaCas9 can direct highly specific genome editing activity and will be useful for targeted genome editing in crops.
Results
Comparing the efficiencies of SaCas9 and SpCas9 for genome editing in tobacco
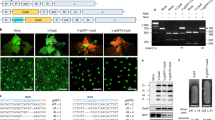
A synthetic SpCas9 gene19 and the SaCas9 gene optimized for Arabidopsis thaliana codon usage were expressed to induce mutation in the tobacco genome. Expression of both Cas9 genes was controlled by the Cauliflower mosaic virus (CaMV) 35S promoter (35S) (Fig. 1a). The gRNA, consisting of a sequence specific to the target DNA and a scaffold sequence, was expressed under the control of the U6-26 promoter20 derived from Arabidopsis thaliana. We selected two target genes in tobacco (Nicotiana tabacum): the PHYTOENE DESATURASE (NtPDS) gene21 and the NtFT4 gene22. Tobacco is an allotetraploid derived from ancestors of two diploids, Nicotiana tomentosiformis and Nicotiana sylvestris, thus there are two homologous genes for PDS and four homologous genes (FT1–4) for FT in the tobacco genome. The PDS gene encodes a phytoene desaturase involving in carotenoid biosynthesis and disruption of PDS causes albino phenotype resulting from lack of carotenoid derivatives such as chlorophyll23. The NtFT4 gene induces flowering in tobacco, while the other three (NtFT1–3) repress flowering22. To evaluate the genome editing activity of SaCas9 in comparison with that of SpCas9, we selected target sequences of 21 nt and 20 nt, the 3′ end of which have 5′-NNGRRT-3′ and 5′-NGG-3′ as the PAM sequences for SaCas9 and SpCas9, respectively (Table 1). To eliminate position effects on genome editing, the two target sequences (SaCas9 and SpCas9) were designed to be mostly same (Table 1). Transgenic tobacco was produced by Agrobacterium-mediated transformation with the constructs shown in Fig. 1a. The transformed tobacco (T0 generation) was regenerated from leaf disks on kanamycin plates for 4–6 weeks (Supplementary Fig. S1a). Genomic DNA was extracted independently from each regenerated shoot and subjected to cleaved amplified polymorphic sequences (CAPS) analysis to assess the presence or absence of mutations in the target sequences. The genome editing frequency was estimated by determining the ratio of regenerated shoots with mutation to those without mutation. The mutation frequency induced by SaCas9 at the NtPDS and NtFT4 gene loci was 75.6% and 65.1%, respectively (Table 2). These results indicate that SaCas9 can induce targeted mutation in tobacco. The mutation frequency induced by SaCas9 was almost same as that induced by SpCas9 at both loci (Table 2). We examined the patterns of mutation induced by SaCas9 and SpCas9 at the NtPDS gene and NtFT4 gene loci (Fig. 1b,c). SaCas9 most often induced small deletions or small insertions around the PAM sequence (Fig. 1b,c). Some transgenic tobacco plants expressing gPDS_Sa and SaCas9 showed an albino phenotype; plants with this phenotype possessed biallelic mutation in two NtPDS genes (Fig. 1d).
SaCas9 induced mutation in tobacco.
(a) Expression construct for SaCas9 (top) and SpCas9 (bottom) in tobacco. A 3 × FLAG tag and 3 × NLS peptide were translationally fused in tandem to the N-terminus of Cas9. The AtADH 5′-UTR and HSP terminator enhance transcription of the Cas9 gene. NLS: nuclear localization signal. AtADH 5′-UTR: 5′ untranslated region of Arabidopsis thaliana ALCOHOL DEHYDROGENASE gene. HSP-ter: the terminator region of Arabidopsis thaliana HEAT SHOCK PROTEIN 18.2 gene. (b,c) Mutations detected in NtPDS (b) and NtFT4 (c) gene loci. The wild-type sequence is shown at the top with the target sequence underlined and the PAM in bold. DNA deletions and insertions are shown in dashes and lower case red letters. The net change in length and the number of clones are noted to the right of each sequence (+, insertion; −, deletion; ×, number of clones). (t): Genomic DNA derived from N. tomentosiformis, (s): genomic DNA derived from N. sylvestris. (d) Phenotype of pds mutant. Left A regenerated plant with wild-type PDS gene, right a regenerated plant with mutation in all four NtPDS genes. (e) Potential off-target genes for NtFT4. The target sequence of the NtFT4 gene is shown at the top with PAM. The mismatched bases in NtFT1 and NtFT2 genes are shown in blue letters. (f) CAPS analysis of NtFT4 gene locus in the T1 generation. Each T1 progeny was segregated from biallelic mutant (T0) or monoallelic mutant (T0). -: Non-digested PCR products, +: DdeI-digested PCR products. An undigested band indicates mutation in the NtFT4 gene.
Analysis of off-target mutations induced by SaCas9
We examined the possibility that SaCas9 induces off-target mutations in tobacco. The NtFT1 and NtFT2 genes have two and four mismatched bases, respectively, in the NtFT4 target sequence (Fig. 1e). No regenerated plants with a mutation in the NtFT1 or NtFT2 gene were detected, suggesting that SaCas9 does not induce mutation in a target sequence that has two mismatched bases in the corresponding gRNA sequence (Table 3).
Heritable targeted mutagenesis using SaCas9
We next examined whether the mutation induced by SaCas9 is inherited by the next generation. Because the pds null mutant is seedling lethal, we analyzed the mutation in the NtFT4 gene. Genomic DNA extracted independently from the T1 progenies of biallelic (T0) and monoallelic (T0) mutant was subjected to CAPS analysis. PCR products that were not digested by restriction enzyme (DdeI) indicated that the mutation had been introduced into NtFT4 gene. All T1 progenies of the T0 biallelic mutant had biallelic mutations induced by SaCas9 in the NtFT4 gene (Fig. 1f). Both biallelic mutants and monoallelic mutants were segregated from the monoallelic mutant in the T1 generation (Fig. 1f). These results indicate that the mutation induced by SaCas9 is inherited to the next generation.
Comparing the efficiency of SaCas9 and SpCas9 for genome editing in rice
The SpCas9 gene24 optimized for rice codon usage was expressed in rice. It has been reported that an Arabidopsis thaliana codon-optimized SpCas9 gene works as well as the Oryza sativa codon-optimized SpCas9 gene in rice25, thus we used Arabidopsis thaliana codon-optimized SaCas9 gene in rice. The expression of both SaCas9 and SpCas9 was controlled by a dual CaMV 35S promoter (2 × 35S) (Fig. 2a). The guide RNA was expressed under the control of the U6-2 promoter derived from Oryza sativa26. We used the DROOPING LEAF (DL) gene for targeted mutagenesis in rice27. The Cas9 gene and gRNA expression vector were both transfected into 1-month-old scutellum-derived rice calli via Agrobacterium-mediated transformation. Transformed rice calli were selected on hygromycin plates for 3 weeks (Supplementary Fig. S1b). The mutation frequency was estimated in two ways as follows: (1) by measuring the ratio of calli with mutation to those without and (2) by measuring the ratio of clones with mutation versus total randomly sequenced clones.
SaCas9 induced mutation in rice.
(a) Expression construct for SaCas9 (upper) and SpCas9 (bottom) in rice. A 3 × FLAG tag and 3 × NLS peptide translationally fused in tandem to the N-terminus of SaCas9. A 1 × FLAG tag was translationally fused to the N-terminus of SpCas9. The OsADH 5′-UTR was added to enhance transcription of the Cas9 gene. OsADH 5′-UTR: 5′ untranslated region of Oryza sativa ALCOHOL DEHYDROGENASE gene. Pea 3A-ter: terminator region of Pea RBCS-3A gene, OsACT1-ter: terminator region of OsACTIN1 gene. (b) CAPS analysis of the OsDL gene locus in SaCas9- and SpCas9-expressing calli. Mutation frequency (%) was calculated from the number of clones with mutation versus total number of sequenced clones indicated under the callus number. -: Non-digested PCR products, +: PstI- or ApaI-digested PCR products. (c) Mutations detected in red rectangle in (b). The wild-type sequence is shown at the top with the target sequence underlined and the PAM in bold. DNA deletions and insertions are shown as dashes and lower case red letters. The net change in length and the number of clones are noted to the right of each sequence (+, insertion; −, deletion; ×, number of clones). (d) Phenotype of dl mutant. Left Wild-type plant, right a regenerated plant from calli with mutation in OsDL gene.
In CAPS analysis, PCR products not digested by restriction enzyme indicated that Cas9 had induced a mutation in the target sequence (Fig. 2b). Mutations were detected in all eight calli expressing gDL-1_Sa and SaCas9 (Fig. 2b, upper left). Also, mutations were detected in all calli expressing gDL-1_Sp and SpCas9 (Fig. 2b, upper right). The same results were obtained in gDL-2_Sa- and SaCas9-expressing calli and gDL-2_Sp- and SpCas9-expressing calli (Fig. 2b, lower). We estimated the mutation frequency in independent calli; PCR products derived from independent calli were cloned into plasmids and sequenced. The mutation frequencies of #1 and #2 callus lines expressing gDL-1_Sa and SaCas9 were 87.5% and 93.7%, respectively. The mutation frequency of #2 callus expressing gDL-1_Sp and SpCas9 was 81.2%. The same results were obtained in gDL-2_Sa, SaCas9- and gDL-2_Sp, SpCas9-expressing callus. These results indicate that SaCas9 could induce mutations in rice just as well as SpCas9 and also that the targeted mutagenesis efficiency of SaCas9 is comparable to that of SpCas9. We examined the patterns of mutation in rice calli caused by SaCas9 and SpCas9 (Fig. 2c). Like SpCas9, small deletions or small insertions close to the PAM sequence were found most often in SaCas9-induced mutations, while large deletions (>25 bp) were rarely detected. We examined the genotype and phenotype of rice plants regenerated from transformed calli that express gDL-1_Sa or gDL-2_Sa and SaCas9 (Fig. 2d and Supplementary Fig. S2). The OsDL gene is a member of the YABBY gene family and the dl mutation causes defects in midrib formation in leaves, resulting in a drooping leaf phenotype28; regenerated rice plants possessing a biallelic mutation in the DL gene exhibited this phenotype.
Base preference for ‘T’ at the 6th position of the SaCas9 PAM in plants
Ran et al. reported that the thymine (T) at the 6th position of the SaCas9 PAM 5′-NNGRRT-3′ is not necessary for genome editing in human cells17. If this is also the case in plants, the selectivity of potential SaCas9 target sequences can be expanded. To explore this possibility, we selected two pairs of target genes (CYP72A33 and OsCYP72A32 and OsVIP1 and OsVIP1-like). Each pair of genes has a perfect-match target sequence differing only at the 6th position of the PAM sequence (Table 1). We applied a heteroduplex mobility assay (HMA) that has been used previously to detect both transcription activator-like effector nucleases (TALENs)-induced mutation29 and CRISPR/Cas9-induced mutation30. Genomic DNA was prepared from individual calli expressing SaCas9 and gCYP72A33_Sa. Each of the PCR products from the CYP72A33 and CYP72A32 gene loci derived from the same genomic DNA was subjected to HMA. The presence of a heteroduplex structure indicates that the targeted mutation has been introduced into the genomic DNA. Heteroduplex structure was detected in the CYP72A33 target sequence that has a ‘T’ at the 6th position of PAM (Fig. 3a upper panel). The number of calli exhibiting the presence of the heteroduplex structure was reduced in the CYP72A32 target sequence, which has a ‘C’ at the 6th position of PAM (Fig. 3a). The frequency of targeted mutagenesis in the CYP72A33 gene was also reduced to 0–4.1% in the CYP72A32 gene. The same results were obtained in the VIP1 gene and VIP1-like gene loci (Fig. 3b). The VIP1 target sequence has a ‘T’ at the 6th position of PAM, while the VIP1-like target sequence has an ‘A’ at this position. We examined the patterns of mutation induced in #4 calli at CYP72A33 and CYP72A32 gene loci (Fig. 3c). We also analyzed the mutation frequency using other target sequences with non-canonical PAM sequences (Table 4). The results suggested that the ‘T’ at the 6th position of PAM is necessary in order for SaCas9 to recognize and cleave the target sequence in plant cells.
Targeted mutagenesis efficiency in target gene with canonical or non-canonical PAM for SaCas9.
(a,b) Heteroduplex mobility assay of SaCas9 expression calli. CYP72A33 and VIP1 target sequences have canonical PAM (5′-NNGRRT-3′) (upper panel). CYP72A32 and VIP1-like target sequences have non-canonical PAM (5′-NNGRRC-3′ and 5′-NNGRRA-3′) (lower panel). (c) Mutations detected in OsCYP72A33 and OsCYP72A32 gene loci. The wild-type sequence is shown at the top with the target sequence underlined and the PAM in bold. DNA deletions and insertions are shown as dashes and lower case red letters. The net change in length and the number of clones are noted to the right of each sequence (+, insertion; −, deletion; ×, number of clones). The asterisk (*) indicates the difference from the canonical PAM for SaCas9.
Discussion
We compared the efficiency of targeted mutagenesis using SaCas9 and SpCas9 on multiple target sequences in rice and tobacco and showed that SaCas9 works with comparable efficiency to SpCas9. Utilization of SaCas9, which recognizes 5′-NNGRRT-3′ as PAM, expands the selectivity of targeted mutagenesis sites in dicots and monocots and will be a useful addition to the widely used SpCas9. We used target sequences with three (5′-NNGAGT-3′, 5′-NNGGGT-3′, 5′-NNGAAT-3′) out of four PAM patterns for SaCas9 at their 3′ end to induce mutations in tobacco and rice. No obvious difference in targeted mutagenesis efficiency was found between them. These findings suggest that both G and A at the 4th or 5th position of PAM for SaCas9 are equally suitable for genome editing in plants.
The efficiency of targeted mutagenesis with non-canonical PAMs (5′-NNGRRV-3′) is much lower than that with a canonical PAM (5′-NNGRRT-3′) in rice (Fig. 3). Ran et al. showed that 5′-NNGRRN-3′ can work as a PAM for SaCas9 in human cells, but that 5′-NNGRRT-3′ was the most efficient in inducing mutation. Kleinstiver et al. showed using a bacterial-based negative selection system that three PAMs (5′-NNGAGT-3′, 5′-NNGGGT-3′, 5′-NNGAAT-3′) were more functional than other PAMs31. These data are consistent with our experimental data in plant cells. We also recommend that 5′-NNGRRT-3′ be used for SaCas9 PAM in plant cells. Therefore, the sequence 5′-NNGRRT-3′ should be selected as the preferred PAM sequence when designing an efficient targeted mutagenesis strategy in plants using the SaCas9 system.
SpCas9 induces off-target mutations, even at the sites that possess five mismatched bases in the corresponding on-target sequence in human cells32,33,34. In rice cells, SpCas9 with gCDKB2, which is targeted to the OsCDKB2 gene, induced off-target mutation in the OsCDKB1 gene, which has two mismatched bases and one of two mismatched bases is located in seed sequence24. NtFT1 gene possesses two mismatched bases at the NtFT4 target sequence for SaCas9 and one mismatched base in the seed sequence (Fig. 1e). However, we detected no mutation in the NtFT1 gene (Table 3). The 21- or 22-nt target sequence for SaCas9 is 1 or 2 nt longer than that of SpCas9 and the PAM for SaCas9 is 2 nt longer than that of SpCas9. We have shown that 5′-NNGRRT-3′ is the preferred PAM for targeted mutagenesis using SaCas9 in plants. These results suggest that the probability of off-target mutation induced by SaCas9 might be lower than that induced by SpCas9. This suggestion is supported by data from Friedland et al.35. Their GUIDE-seq result shows that SaCas9 is more specific than SpCas9 in human cells35.
Methods
Plasmid construction
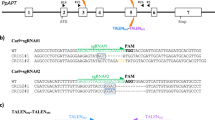
The Arabidopsis thaliana codon-optimized SaCas9 gene was synthesized by GeneArt Gene Synthesis (Thermo Fisher Scientific, USA). The backbone of the binary vector used in this study was derived from pRI201-AN (TaKaRa, Japan) and pPZP200 for tobacco and rice, respectively. The Cas9 expression vector in tobacco, pRI201-AN was constructed as follows: (1) the Arabidopsis thaliana codon-optimized Cas9 was fused to 3 × FLAG tag (Sigma-Aldrich, USA) and 3 × NLS (nuclear localization signal), respectively. (2) CaMV 35S promoter (35S) and the sequences of Arabidopsis thaliana alcohol dehydrogenase (ADH) 5′-untranslated region (UTR) were connected upstream of Cas9 and the terminator sequence of Arabidopsis thaliana HEAT SHOCK PROTEIN (HSP) gene was connected downstream of Cas9 (Supplementary Figs S3 and S4). The Cas9 expression vector in rice, pPZP200, was constructed as follows: (1-1) the Arabidopsis thaliana codon-optimized SaCas9 was fused to 3 × FLAG tag and 3 × NLS. (1–2) the Oryza sativa codon-optimized SpCas9 was fused to 1 × NLS. (2) the dual CaMV 35S promoter (2 × 35S) and the sequences of the rice alcohol dehydrogenase (ADH) 5′-untranslated region (UTR) were connected upstream of Cas9 and the pea RBCS3A (pea3A) terminator sequence and the sequence of the Oryza sativa ACTIN1 (ACT1) gene 3′-UTR were connected downstream of Cas9 (Supplementary Figs S5 and S6). The guide RNA expression vector was constructed as follows. (1) The sequence of the OsU6 promoter and AtU6, RNA scaffold and poly (T) were synthesized. (2) The synthesized target sequence (21 nt or 20 nt) was cloned into the BbsI site of the single guide RNA expression vector (Supplementary Fig. S7). The PacI-AscI fragment of the single guide RNA expression vector was subcloned into the Cas9 expression binary vector.
Transformation of rice callus
Agrobacterium-mediated transformation of rice (Oryza sativa L. cv. Nipponbare) using scutellum-derived calli was performed as described previously36,37. Briefly, one-month-cultured rice calli were infected by Agrobacterium (EHA105 strain). After 3 days of co-cultivation, calli were transferred to callus-induction medium containing 50 mg/L hygromycin B (Wako Pure Chemicals, Osaka, Japan) and 25 mg/L meropenem (Wako Pure Chemicals, Japan). Hygromycin-resistant calli were selected over 4 weeks.
Transformation of tobacco
Tobacco (Nicotiana tabacum L. cv. Petit Havana SR-1) was transformed via Agrobacterium-mediated transformation as described previously38. Transgenic tobacco plants were regenerated from leaf disks on medium containing 50 mg/L kanamycin (Wako Pure Chemicals, Japan) and 25 mg/L meropenem.
CAPS analysis
Genomic DNA was extracted from calli, regenerated rice plants or transgenic tobacco shoots using Agencourt Chloro Pure (BECKMAN COULTER, USA) and target loci were amplified by PCR. PCR products were digested by restriction enzymes.
Heteroduplex mobility assay
Heteroduplex mobility assay (HMA) was applied to detect mutations induced by Cas9 with a microchip electrophoresis system30. PCR products were analyzed using MCE-202 MultiNA with a DNA-500 kit (SHIMADZU, Japan).
Additional Information
Accession codes: NtFT4; AWOKO1087688, DROOPING LEAF; Os03g0215200/LOC_Os03g11600 LOC_Os11g04954, CYP72A32; LOC_Os01g41810, CYP72A33; LOC_Os01g41820, VIP1; LOC_Os12g06520, VIP1-like; LOC_Os11g06170, OsALS; LOC_Os02g30630, OsLIG6; Os01g49180, OsPDS; LOC_Os03g08570, OsPOLQ; LOC_Os12g19370. http://www.nature.com/srep
How to cite this article: Kaya, H. et al. Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci. Rep. 6, 26871; doi: 10.1038/srep26871 (2016).
References
Bhaya, D., Davison, M. & Barrangou, R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45, 273–297, doi: 10.1146/annurev-genet-110410-132430 (2011).
Jiang, W. & Marraffini, L. A. CRISPR-Cas: New Tools for Genetic Manipulations from Bacterial Immunity Systems. Annu Rev Microbiol 69, 209–228, doi: 10.1146/annurev-micro-091014-104441 (2015).
Doudna, J. A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096, doi: 10.1126/science.1258096 (2014).
Carroll, D. Genome engineering with targetable nucleases. Annu Rev Biochem 83, 409–439, doi: 10.1146/annurev-biochem-060713-035418 (2014).
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N. J. & Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol 32, 76–84, doi: 10.1016/j.copbio.2014.11.007 (2015).
Osakabe, Y. & Osakabe, K. Genome editing with engineered nucleases in plants. Plant Cell Physiol 56, 389–400, doi: 10.1093/pcp/pcu170 (2015).
Baltes, N. J. & Voytas, D. F. Enabling plant synthetic biology through genome engineering. Trends Biotechnol 33, 120–131, doi: 10.1016/j.tibtech.2014.11.008 (2015).
Schaeffer, S. M. & Nakata, P. A. CRISPR/Cas9-mediated genome editing and gene replacement in plants: Transitioning from lab to field. Plant Sci 240, 130–142, doi: 10.1016/j.plantsci.2015.09.011 (2015).
Bortesi, L. & Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33, 41–52, doi: 10.1016/j.biotechadv.2014.12.006 (2015).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823, doi: 10.1126/science.1231143 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826, doi: 10.1126/science.1232033 (2013).
Deltcheva, E. et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607, doi: 10.1038/nature09886 (2011).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821, doi: 10.1126/science.1225829 (2012).
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67, doi: 10.1038/nature13011 (2014).
Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J. & Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740, doi: 10.1099/mic.0.023960-0 (2009).
Fonfara, I. et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res 42, 2577–2590, doi: 10.1093/nar/gkt1074 (2014).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191, doi: 10.1038/nature14299 (2015).
Steinert, J., Schiml, S., Fauser, F. & Puchta, H. Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J, doi: 10.1111/tpj.13078 (2015).
Fauser, F., Schiml, S. & Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79, 348–359, doi: 10.1111/tpj.12554 (2014).
Li, X., Jiang, D. H., Yong, K. L. & Zhang, D. B. Varied transcriptional efficiencies of multiple Arabidopsis U6 small nuclear RNA genes. J Integr Plant Biol 49, 222–229, doi: 10.1111/j.1672-9072.2007.00393.x (2007).
Busch, M., Seuter, A. & Hain, R. Functional analysis of the early steps of carotenoid biosynthesis in tobacco. Plant Physiol 128, 439–453, doi: 10.1104/pp.010573 (2002).
Harig, L. et al. Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J 72, 908–921, doi: 10.1111/j.1365-313X.2012.05125.x (2012).
Qin, G. et al. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid and gibberellin biosynthesis. Cell Res 17, 471–482, doi: 10.1038/cr.2007.40 (2007).
Endo, M., Mikami, M. & Toki, S. Multigene Knockout Utilizing Off-Target Mutations of the CRISPR/Cas9 System in Rice. Plant Cell Physiol 56, 41–47, doi: 10.1093/pcp/pcu154 (2015).
Mikami, M., Toki, S. & Endo, M. Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88, 561–572, doi: 10.1007/s11103-015-0342-x (2015).
Feng, Z. Y. et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23, 1229–1232, doi: 10.1038/cr.2013.114 (2013).
Yamaguchi, T. et al. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16, 500–509, doi: 10.1105/tpc.018044 (2004).
Nagasawa, N. et al. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718 (2003).
Ota, S. et al. Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells 18, 450–458, doi: 10.1111/gtc.12050 (2013).
Ansai, S. & Kinoshita, M. Targeted mutagenesis using CRISPR/Cas system in medaka. Biol Open 3, 362–371, doi: 10.1242/bio.20148177 (2014).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485, doi: 10.1038/nature14592 (2015).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31, 822–826, doi: 10.1038/nbt.2623 (2013).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–832, doi: 10.1038/nbt.2647 (2013).
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31, 839–843, doi: 10.1038/nbt.2673 (2013).
Friedland, A. E. et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol 16, 257, doi: 10.1186/s13059-015-0817-8 (2015).
Toki, S. Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol Biol Rep 15, 16–21, doi: 10.1007/Bf02772109 (1997).
Toki, S. et al. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant Journal 47, 969–976, doi: 10.1111/j.1365-313X.2006.02836.x (2006).
Taoka, K. et al. Identification of three kinds of mutually related composite elements conferring S phase-specific transcriptional activation. Plant Journal 18, 611–623, doi: 10.1046/j.1365-313x.1999.00486.x (1999).
Acknowledgements
We thank Dr. H. Puchta for kindly providing the Arabidopsis thaliana codon-optimized SpCas9 gene, Drs. H. Nishimasu and O. Nureki for providing technical information, Drs. A. Nishizawa-Yokoi, H. Saika, S. Hirose, K. Abe and N. Ohtsuki for advice and discussion and K. Amagai, R. Aoto, C. Furusawa, A. Mori, A. Nagashii, A. Nakano, F. Suzuki and R. Takahashi for general experimental technical support. This work was supported by the Cross-ministerial Strategic Innovation Promotion Program (SIP).
Author information
Authors and Affiliations
Contributions
H.K., M.M. and S.T. designed the experiments. H.K. and M.M. performed all experiments. All authors analyzed the results and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kaya, H., Mikami, M., Endo, A. et al. Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci Rep 6, 26871 (2016). https://doi.org/10.1038/srep26871
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26871
This article is cited by
-
CRISPR/Cas system for the traits enhancement in potato (Solanum tuberosum L.): present status and future prospectives
Journal of Plant Biochemistry and Biotechnology (2024)
-
Enhancing the quality of staple food crops through CRISPR/Cas-mediated site-directed mutagenesis
Planta (2023)
-
Efficiency verification of CRISPR–Cas9-mediated mutagenesis of target gene sgRNA using soybean protoplasts
Plant Biotechnology Reports (2022)
-
PAM-less plant genome editing using a CRISPR–SpRY toolbox
Nature Plants (2021)
-
Expanding the range of CRISPR/Cas9-directed genome editing in soybean
aBIOTECH (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.