Abstract
While single nucleotide polymorphism (SNP) is typically the variant of choice for population genetics, copy number variation (CNV) which comprises insertion, deletion and duplication of genomic sequence, is an informative type of genetic variation. CNVs have been shown to be both common in mammals and important for understanding the relationship between genotype and phenotype. However, CNV differentiation, selection and its population genetic properties are not well understood across diverse populations. We performed a population genetics survey based on CNVs derived from the BovineHD SNP array data of eight distinct cattle breeds. We generated high resolution results that show geographical patterns of variations and genome-wide admixture proportions within and among breeds. Similar to the previous SNP-based studies, our CNV-based results displayed a strong correlation of population structure and geographical location. By conducting three pairwise comparisons among European taurine, African taurine, and indicine groups, we further identified 78 unique CNV regions that were highly differentiated, some of which might be due to selection. These CNV regions overlapped with genes involved in traits related to parasite resistance, immunity response, body size, fertility, and milk production. Our results characterize CNV diversity among cattle populations and provide a list of lineage-differentiated CNVs.
Similar content being viewed by others
Introduction
Copy number variations (CNVs) are large-scale insertions and deletions, existing as one type of complex multiallelic variants within diverse populations1,2. Compared to single nucleotide polymorphisms (SNPs), CNVs involve more genomic sequences and have potentially greater effects, including changing gene structure and dosage, altering gene regulation and exposing recessive alleles3. Human and mouse studies found that CNVs captured 18–30% of the genetic variation in gene expression4,5. These CNVs were shown to be important in both normal phenotypic variability and disease susceptibility. Population genetics has played an important role in exploring genetic variations in human6 and farm animals7. Investigating the population genetics and evolutionary origins of CNVs could enable us to understand their origins and impacts8,9,10,11. With recent advances in our knowledge of the locations, sizes and mutational mechanisms of CNV using high-throughput screening approaches, the attempt to study corresponding population genetics is gradually developing in human and other model species. Findings from these initial studies have brought new insights into genome diversity and adaptation12,13,14,15.
Population structure analyses based on human CNVs have revealed results largely consistent with those based on SNPs of similar number16. For instance, based on hybrid genotyping arrays, up to 90% of human CNVs can be revealed by integrated investigation of SNPs17. On the other hand, multiple lines of evidence also suggest CNVs could serve as an extra genomic resource and provide important insights into the origins and sub-structure of populations9,15,16,18,19,20,21,22. Additionally, population-specific CNVs are candidate regions under selection and are potentially responsible for diverse phenotypes9,23,24.
Previous studies have also revealed that genomic diversity could be generated by the bias of selection on CNV in specific environments for adaptations25. For instance in human adaptations, positive selection for a higher AMY1 copy number enables the better digestion of starchy foods26. An indel polymorphism in gene APOBEC3b has been associated with malaria susceptibility27. The human UGT2B17 gene shows significant copy-number diversity among populations from Africa, Europe, and East Asia, which displays region-specific differences in the metabolism of steroid hormones and a large number of xenobiotics28. Another well-known example is the olfactory receptor (OR) genes, which are frequently found to be copy-number variable in most mammalian species. The differences in OR gene counts between human populations suggested that they are involved in population-specific differences in smell29. In addition, CNVs are specifically enriched among evolutionary “young” ORs, implying that CNVs may play a critical role in the processes of gene birth and death or the emergence of new OR gene clusters30.
In livestock, such as cattle, most CNV studies have limited themselves to CNV detection and enumeration using various platforms, such as CGH array, SNP array or next generation sequencing31,32,33,34,35,36,37,38,39. Even though the aforementioned studies have identified a large number of copy number variable regions in their respective species, exploring livestock population genetics using cattle CNVs is still in its infancy. The investigation of diversity and origin of CNVs, the characterization of their population-genetic properties, and the determination of the functional impacts of CNVs are still active areas of research.
Here, we report a comprehensive population-genetics study of CNVs by focusing on the diversity, population structure, and selection of identified CNVs within eight representative cattle breeds. In this study, we investigated CNVs from individuals originating from European taurine, indicine, and African taurine breeds of the Bovine HapMap DNA panel40. Our results revealed that most common CNVs, especially CNV deletions, show large differences in frequency across diverse groups. More importantly, we demonstrated that CNVs can be used for the investigation of population genetics in cattle, as we observed CNVs with significant diversity across groups that might be associated with breed and sub-species specific selection signatures.
Results
CNVs segmentation and genotyping
A total of 300 individuals was used for CNV discovery as shown in Table S1, including Holstein (HOL), Angus (ANG), Hereford (HFD), Brown Swiss (BWS), Brahman (BRM), Nelore (NEL), N’Dama (NDA), and Sheko (SHK). In total, 155,700 CNV segments were extracted by Golden Helix SVS 8.0 using the default multivariate option. After merging across all individuals, we discovered 263 non-redundant CNVs which are commonly shared within the whole population (Table S2). Since the SVS multivariate option was developed to identify moderate to high frequency CNVs, only segments with frequencies above 1% were retained for further analysis in order to filter away potential false positive calls. Finally, a total of 257 CNVs (with a total length of 12,444 kb and an average length of 48.4 kb) were retained and used to categorize the samples as one of three types (loss, neutral and gain events) according to a three-state model with strict threshold levels of marker mean log R ratio (LRR) ± 0.3. They were sorted as a list of CNV1 to CNV257 with a descending frequency, in which there were 184 deletion CNVs (Table S2). As shown previously41,42, comparisons of CNV detection algorithms usually revealed a low concordance. However, when we compared this dataset with our previous results using PennCNV in the same Bovine Hap Map samples32, we obtained a total of 160 concordant CNVs (61%), indicating a high quality of our SVS results. While all 257 CNVs were used for frequency and VST calculations, only the 184 deletion CNV regions were used in all other subsequent population genetics analyses.
Population-genetic properties of cattle CNVs
Hierarchical Clustering Analysis
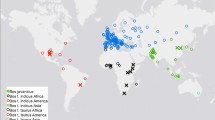
To obtain a global picture of group differences, hierarchical clustering was done using the mean LRRs for the 257 CNVs. Three distinct groups were observed, including group one European taurine (TAU) containing HOL, ANG, HFD, and BWS; a second indicine (IND) group containing BRM and NEL; and third group African taurine (AFR) containing NDA (Fig. 1). SHK, which used to be considered a taurine population, because they are humpless, was positioned between IND and AFR confirming SHK is a hybrid breed in agreement with its known breed formation history43.
Multidimentional Scaling (MDS) Analysis
To examine the population structure of these three cattle groups based on CNVs, a multidimensional scaling (MDS) analysis was completed on 205 unrelated individuals based on the 184 deletion CNVs (Fig. 2A). We found C1 axis can clearly separate taurine (TAU and AFR) from indicine (IND), while C2 axis can separate African taurine (AFR) into its unique cluster with a small amount of intermixing with European taurine (TAU). Therefore, the global organization of cattle genetic diversity can be represented as a triangle with apexes corresponding respectively to TAU, IND, and AFR groups. As expected, we observed that SHK was located between AFR and IND, again confirming its hybrid breed formation history. We found this CNV-based MDS results are generally consistent with the results from a similar SNP-based analysis40,44, suggesting CNVs can be used to separate cattle individuals into distinct groups. However, the clustering resolution within groups based on CNVs was not better than those based on SNPs. For example, CNVs cannot distinguish the HOL breed from the ANG breed in European taurine cattle. There were also certain degrees of mixing within indicine individuals in the CNV-based clustering results (Fig. 2A). In summary, our results revealed that CNV can be used in population genetic studies. However, compared to SNP, CNV suffers from small sampling size and difficulty to genotype, making it difficult to use them to do fine clustering, especially within a group.
Four distinct groups include the European taurine (TAU) group containing HOL, ANG, HFD, and BWS; the second indicine (IND) group containing BRM and NEL; the third group African taurine (AFR) containing NDA; and the fourth group formed by the hybrid SHK. (A) MDS analysis of 205 individuals. Individuals were plotted according to their coordinates on the first two components. (B) Clustering of 205 individuals from eight breeds based on 184 deletion CNVs when K = 2–6. Individuals were shown as a thin vertical line colored in proportion to their estimated ancestry. (C) Neighbor-joining tree of the 205 individuals. The tree was constructed using genetic sharing distances. Edges were labeled according to the breed of origin.
Admixture Analysis
To investigate genome wide ancestral admixture patterns of these eight breeds, we used the admixture inference method implemented in STRUCTURE (Fig. 2B). Varying the number of presumed ancestral populations (K) recapitulated the extent of genetic divergences across breeds. At K = 2, TAU and AFR were clearly assigned into unique groups distinct from IND. At K = 3, the clustering analysis revealed TAU was separated from AFR showing a clear separation of TAU, IND and AFR groups. At K = 4, intriguingly, European taurine beef breeds was separated from their dairy counterparts. At K = 5, BWS was deviated from HOL. Finally at K = 6, HFD was separated from ANG and most of the samples were clustered according to breed designation, except that the NEL and BRM breeds were still clustered together. In addition, increasing the number of inferred clusters allowed us to confirm a high level of admixture and support the documented origin of SHK, which accommodated high fractions of admixture from ancestries of AFR and IND. Overall, these results were in agreement with our MDS analysis, suggesting that the partitioning of cattle into distinct populations is closely related to genetic diversity, which is in agreement with the earlier report by Bovine HapMap Consortium40.
Neighbor-Joining Clustering Analysis
In addition, we calculated all pairwise genetic distances using PLINK 1.07, and plotted a neighbor-joining dendrogram of all individuals (Fig. 2C). We found that the genetic relationship among cattle groups could be largely recovered from this dendrogram as it clearly arranged individuals according to their population of origin. Although two indicine breeds (BRM and NEL) are intermixed, the three breed groups (TAU, AFR and IND) can be easily distinguished. In agreement with MDS and admixture results, individuals from SHK branched between AFR and IND. This clustering analysis of individual samples supports most of the relationships among the cattle breeds uncovered by our MDS and STRUCTURE analysis.
Population diversity and differentiated CNVs
Using 257 CNVs, we estimated the CNV frequencies across 3 cattle groups, i.e. TAU, IND and AFR. With an average of frequency of 0.39, we indentified the top five high frequency deletions were on chromosomes 11, 26, 4, 29, and 11 with the corresponding frequencies of 0.93, 0.93, 0.92, 0.92, and 0.90, respectively (Table S2). Using the 205 unrelated individuals, we estimated the frequencies for each group to investigate CNVs with differentiated frequencies.
We compared CNV regions with the UMD 3.1 Ensembl gene (EnsGene) annotation and found 101 CNVs partially overlapping with genes (Table S3). We performed gene ontology (GO) analysis using PANTHER45 to identify if there was enrichment of genes with specific function (Table S4). The most enriched biological processes include response to interferon-gamma (Interferon-Induced Guanylate-Binding Protein 2), other immunity related processes, and response to stimulus (MHC, immunoglobins, ORs and ATP-binding cassette (ABC) transporters). Aside from 99 CNVs that overlapped with EnsGene genes, we found 158 CNVs which did not encompass any EnsGene genes. When comparing CNVs overlapped with genes to CNVs not overlapped with genes, we found that the CNVs without genes were shorter (with Mean 23370 ± Standard Error of Mean 5659, N = 158 vs. 88400 ± 18500, N = 99, t-test, p-value < 0.0001) and at higher frequency (0.4218 ± 0.0189, N = 158 vs. 0.3468 ± 0.0218, N = 99, p-value = 0.0113). The paucity of common deletion CNVs overlapping with genes is consistent with the notion that they are under purifying selection, which removes deleterious variants from the population. We noted that our length and frequency analysis could be confounded if the power to detect short events is higher than long events and the power to detect common deletions is higher than common duplications. Besides other neutral possibilities, like those indicated in an early human study14, CNVs may also affect gene expression through regulatory level changes.
CNVs that differ greatly in frequency between cattle groups/breeds are candidates for population-specific selection. To test whether any CNV might be associated with population-specific selection, we estimated the pairwise VST for 4 comparisons, including TAU vs. IND, TAU vs. AFR, IND vs. AFR, and HOL vs. ANG (Fig. 3, Fig. S1, and Table S3). VST estimations produce values from 0 (no difference) to 1 (complete population differentiation), with high VST values indicating regions under increased selective pressure or other evolutionary forces, such as bottlenecks or founder effects. Using a stringent threshold cutoff of VST > 0.6, we detected a total of 14, 0, 18, 1 CNV(s) in the abovementioned four comparisons, respectively. When we lowered the threshold to 0.4, we observed 41, 11, 48, 2 CNVs for the four comparisons, respectively.
The higher differential VST identified in these CNV regions may suggest the dosage variability of their underlying genomic sequence, which could be further involved in the diverse phenotypes across cattle breeds. For instance, when comparing TAU with IND under the lower threshold of 0.4, we observed ten genes overlapped with CNV regions, including CDH18, GDAP1L1, HIATL1, IGLL1, ITGB8, KCNIP3, LCT, NETO1, OIT3, and SHISA9. Similarly for the comparisons of TAU vs. AFR and IND vs. AFR, we found nine genes (EPHB3, FANCC, GRM7, HSFY2, KCNJ12, LIPF, PRAME, TSPY, and ZNF280B) and nine genes (GDAP1L1, HIATL1, LCT, MRPL48, MSMB, PLCB1, RBFOX1, ROBO4, and SHISA9) overlapping with CNV regions, respectively. Although for some genes, only small parts were covered by CNVs, the change of these small regions could potentially influence their function and evolution. We further overlapped these genes with the known cattle quantitative trait locus (QTL at http://www.animalgenome.org/cgi-bin/QTLdb/BT/index) and found genes associated with important traits in cattle that vary in copy number frequencies across populations (Fig. S2). For example, some of them are related to parasite resistance, immunity response and adaption, including EPHB3, SHISA9 and LCT46,47. Other genes were reported to be involved in body size, fertility, production and milk fatty acid profile, for examples FANCC, IGLL1 and LIPF48,49.
Discussion
Population genetics studies based on CNVs have been explored in human, dog, zebrafish, and stickleback fish8,9,23,50. Despite that previous studies have identified CNVs within and between populations in cattle, our study is one of the first attempts to explore the population-genetic properties in cattle based on CNVs derived from the high-density SNP array. We also provided additional evidence to support CNVs as genetic markers that can be used to study the across population diversity and capture the subspecies relationships. Since it was difficult to accurately detect and genotype complex CNV events, like non-biallelic duplications, in this proof-of-principle study, we mainly used high confidence deletion CNVs. The distinct advantage of deletion CNVs over duplication events is that deletions can be treated as bi-allelic markers, and are therefore compatible with mature genetics analysis methods designed for SNP markers.
In the current study, we used CNVs with moderate intra-population frequencies to explore the population-genetic properties in cattle. Although the SVS method we utilized reported a limited amount of CNV calls, including 71.6% deletions and 28.4% duplications, our study did reveal that globally diverse cattle populations clustered roughly by geographical region and were influenced by demographic history, which is similar to the results derived from previous SNP-based cattle studies40,51,52. This could be due to the fact that 75% of the CNVs that we identified were well tagged by flanking SNPs, as was estimated previously53. We also found the results of this survey were not capable of distinguishing recently divergent cattle breeds of common geographic origin, such as HOL and ANG breeds, which were reported to be separated by 364 generations54. This observation was consistent with human studies, where published reports also showed the CNV-based population stratification can be only detected among the large population groups at the continent level15. The fine genetic structure detected by SNPs may be due to their accuracy of genotyping and sample size, besides other influencing factors, as suggested previously16. Further study based on high throughput sequencing will make it easier to accurately genotype CNVs11,34.
To investigate lineage-differentiated CNVs in the cattle genome, we also conducted CNV-based population differentiation analysis, and identified potential CNV candidates under divergent selection. We estimated VST values, a population differentiation estimator similar to FST, among groups and examined gene enrichments among CNVs regions. In our previous study based on array CGH in cattle populations, we revealed that regions that have been under recent positive selection exhibit elevated population differentiation33. In the current study, we found 78 unique CNVs with VST values above the threshold of 0.4 as potential lineage- differentiated events in three group comparisons, perhaps representing increased selective pressures exerted upon the cattle population. It is noted that besides selective pressure, the amount of divergence between populations (time since divergence, effective population size, and gene flow/migration) also can affect the overall differentiation and VST values. High VST between groups does not necessarily involve divergent selection (selection in both populations for different alleles), and can also occur in the absence of selection, for example, by bottlenecks or founder effects followed by drift. All these hypotheses warrant further investigations using larger sample sizes.
By contrast, we observed only a handful of lineage-differentiated CNVs in our HOL vs. ANG comparison, which did not overlap with any known cattle genes. Fewer lineage-differentiated CNVs may suggest that these two breeds might not have had sufficient time to diversify their deletion CNV contents, if we assume that the CNV occurred before the split of the two breeds, and that the deletion event should eventually fix in one but not the other population. Additionally, fewer lineage-differentiated CNVs were also observed in laboratory mouse and zebrafish strains, as compared with their wild populations23,55. This may be indicative of either inbreeding effects or suggest that CNVs were preferentially fixed as a consequence of the larger effective population size of wild populations. It is well known that HOL and ANG have gone through intensive human selection recently via the practice of artificial insemination.
In the future, we propose that additional cattle breeds from places like the Middle East, Pakistan and Turkey will provide more insights into the worldwide and local genetic diversity and population structure. We also expect that discovery of low frequency CNV variants, especially in those under-represented breeds like indicine and African taurine cattle will provide additional resolution for distinguishing those populations. Finally, with more powerful software tools56, we predict population genetics in livestock will remarkably expand with next generation sequencing data.
Methods
Samples
In the CNV discovery phase, we retrieved a subset of Illumina BovineHD SNP dataset (300 individuals, Table S1), which represent 8 geographically diverse breeds, including Holstein, Angus, Hereford, Brown Swiss, N’Dama, Sheko, Brahman, and Nelore32. All chosen samples had a genotyping success rate of more than 99%. For population genetic analyses, we only used 205 animals after removing related individuals according to pedigree information and pi-hat value if it was more than 0.4.
CNV segmentation and genotyping
The intensity data of 742,910 SNP probes were generated using the Illumina BovineHD SNP array. After exporting the DSF file from GenomeStudio Software, we imported Log R Ratio (LRR) into Golden Helix SNP & Variation Suite (SVS) 8.0 (Golden Helix Inc., Bozeman, MT, USA) and successfully mapped 735,293 SNPs (98.97%) onto the 29 autosomes of Bos taurus genome assembly UMD 3.1. The LRR was then normalized using the default GC correlation file to correct the waviness caused by the GC content. We then utilized the copy number analysis module (CNAM) under the multivariate option to segment chromosomes with default set, and a significance level of p = 0.01 for pairwise permutations (n = 1,000) as described previously53. The three state covariates with a comparatively strict threshold (segment mean 0.3) was used to genotype the CNVs as one of three type (loss, neutral and gain events) across all the samples. It was noted that the multivariate method tends to detect the common deletions with relative small sizes across multiple samples.
Population differences across population
We first checked the normalized LRR distribution histograms for all 184 deletion CNVs. The grand majority (99%) of deletions had two distinct peaks, representing neutral (around 0) and homozygous deletion (around -1) states, respectively. Only a couple of deletions had a few samples located in the midpoint between -1 and 0, suggesting a lack of heterozygous events. Additionally, it was difficult to define a universal threshold between homo or heterozygous deletions for all deletions, therefore, we decided to categorize all deletion events using one state: homozygous deletion. To use population genetic programs originally developed for SNPs, we manually recoded each 184 deletion CNVs by converting a loss event into “12” or a neutral event into “22”, where “12” represented a homozygous deletion.
The R Function heatmap.2 (http://www.inside-r.org/packages/cran/gplots/docs/heatmap.2) was used to graph the segment mean LRR values and generate hierarchical cluster dendrograms using 257 CNVs for all animals. We then performed multidimensional scaling (MDS) and admixture analysis to determine how 205 unrelated individuals were clustered according to these CNV genotypes. Using a total of 184 deletion CNVs, MDS analysis of pairwise genetic distance (4 dimensions) was used to detect the relationship between populations with PLINK 1.07 (-mds -plot 4). For a separate verification, we also performed the cluster analysis based on mean LRR values using prcomp functions in R v13.1, the results were consistent with MDS analysis.
Population structure was examined using STRUCTURE 2.357,58. Each admixture analysis was performed using 5,000 replicates and 2,000 burn-in cycles under admixture and allele frequencies correlated models.
Neighbor-joining clustering analysis were performed using PHYLIP 3.69 (http://www.phylip.com/) based on pairwise genetic distance. Pairwise genetic distance (D) between individuals was calculated using PLINK 1.07, where D = 1-[IBS2 + 0.5IBS1)/N], and IBS2 and IBS1 are the number of loci that share either 2 or 1 alleles identical by state (IBS), respectively and the N is the number of loci59,60. The clustering dendrograms were plotted in Figtree 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Gene annotation and PANTHER analysis
We retrieved RefSeq, Ensembl, and xenoRefSeq genes overlapping CNV regions by at least 1 bp, including the 5′ and 3′ untranslated regions, from available UCSC genome browser tracks and annotated CNV regions using custom software (https://github.com/njdbickhart/AnnotateUsingGenomicInfo). We performed enrichment analysis using PANTHER classification system45. Only clusters with enrichment scores more than 1 (p-value < 0.05 after the Bonferroni correction for multiple testing) were considered.
Signatures of Selection
To detect the lineage differentiated CNV events, we calculated VST for each CNV as previously described15 by using the following equation: (VT −VS)/VT, where VT is the total variance in mean LRRs across all individuals and VS is the average variance in cattle within each breed.
Additional Information
How to cite this article: Xu, L. et al. Population-genetic properties of differentiated copy number variations in cattle. Sci. Rep. 6, 23161; doi: 10.1038/srep23161 (2016).
References
Scherer, S. W. et al. Challenges and standards in integrating surveys of structural variation. Nat Genet 39, S7–15 (2007).
Mills, R. E. et al. Mapping copy number variation by population-scale genome sequencing. Nature 470, 59–65 (2011).
Zhang, F., Gu, W., Hurles, M. E. & Lupski, J. R. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 10, 451–481 (2009).
Stranger, B. E. et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315, 848–853 (2007).
Orozco, L. D. et al. Copy number variation influences gene expression and metabolic traits in mice. Hum Mol Genet 18, 4118–4129 (2009).
Lachance, J. & Tishkoff, S. A. Population Genomics of Human Adaptation. Annu Rev Ecol Evol Syst 44, 123–143 (2013).
Larson, G. & Burger, J. A population genetics view of animal domestication. Trends Genet 29, 197–205 (2013).
Sudmant, P. H. et al. Diversity of Human Copy Number Variation and Multicopy Genes. Science 330, 641–646 (2010).
Berglund, J. et al. Novel origins of copy number variation in the dog genome. Genome Biol 13, R73 (2012).
Sjodin, P. & Jakobsson, M. Population genetic nature of copy number variation. Methods Mol Biol 838, 209–223 (2012).
Pronold, M., Vali, M., Pique-Regi, R. & Asgharzadeh, S. Copy number variation signature to predict human ancestry. BMC Bioinformatics 13, 336 (2012).
Conrad, D. F. & Hurles, M. E. The population genetics of structural variation. Nat Genet 39, S30–S36 (2007).
Freeman, J. L. et al. Copy number variation: new insights in genome diversity. Genome Res 16, 949–961 (2006).
Conrad, D. F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010).
Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006).
Jakobsson, M. et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451, 998–1003 (2008).
McCarroll, S. A. et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 40, 1166–1174 (2008).
Kato, M. et al. Population-genetic nature of copy number variations in the human genome. Hum Mol Genet 19, 761–773 (2010).
Campbell, C. D. et al. Population-genetic properties of differentiated human copy-number polymorphisms. Am J Hum Genet 88, 317–332 (2011).
Lou, H. et al. A map of copy number variations in Chinese populations. PLoS One 6, e27341 (2011).
Narang, A. et al. Extensive copy number variations in admixed Indian population of African ancestry: potential involvement in adaptation. Genome Biol Evol 6, 3171–3181 (2014).
Xu, L. et al. Genomic signatures reveal new evidences for selection of important traits in domestic cattle. Mol Biol Evol 32, 711–725 (2015).
Brown, K. H. et al. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci USA 109, 529–534 (2012).
Gautam, P. et al. Spectrum of large copy number variations in 26 diverse Indian populations: potential involvement in phenotypic diversity. Hum Genet 131, 131–143 (2012).
Iskow, R. C., Gokcumen, O. & Lee, C. Exploring the role of copy number variants in human adaptation. Trends Genet 28, 245–257 (2012).
Perry, G. H. et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet 39, 1256–1260 (2007).
Jha, P. et al. Deletion of the APOBEC3B gene strongly impacts susceptibility to falciparum malaria. Infect Genet Evol 12, 142–148 (2012).
Xue, Y. et al. Adaptive evolution of UGT2B17 copy-number variation. Am J Hum Genet 83, 337–346 (2008).
Waszak, S. M. et al. Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput Biol 6, e1000988 (2010).
Hasin, Y. et al. High-resolution copy-number variation map reflects human olfactory receptor diversity and evolution. PLoS Genet 4, e1000249 (2008).
Jiang, L. et al. Genome-wide identification of copy number variations in Chinese Holstein. PLoS ONE 7, e48732 (2012).
Hou, Y. et al. Fine mapping of copy number variations on two cattle genome assemblies using high density SNP array. BMC Genomics 13, 376 (2012).
Liu, G. E. et al. Analysis of copy number variations among diverse cattle breeds. Genome Res 20, 693–703 (2010).
Bickhart, D. M. et al. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res 22, 778–790 (2012).
Fadista, J., Thomsen, B., Holm, L. E. & Bendixen, C. Copy number variation in the bovine genome. BMC Genomics 11, 284 (2010).
Bae, J. S. et al. Identification of copy number variations and common deletion polymorphisms in cattle. BMC Genomics 11, 232 (2010).
Seroussi, E. et al. Analysis of copy loss and gain variations in Holstein cattle autosomes using BeadChip SNPs. BMC Genomics 11, 673 (2010).
Hou, Y. et al. Genomic characteristics of cattle copy number variations. BMC Genomics 12, 127 (2011).
Cicconardi, F. et al. Massive screening of copy number population-scale variation in Bos taurus genome. BMC Genomics 14, 124 (2013).
The Bovine HapMap Consortium. Genome wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324, 528–532 (2009).
Xu, L., Hou, Y., Bickhart, D. M., Song, J. & Liu, G. E. Comparative analysis of CNV calling algorithms: literature survey and a case study using bovine high-density SNP data. Microarrays 2, 171–185 (2013).
Pinto, D. et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol 29, 512–U76 (2011).
Gautier, M. et al. A whole genome Bayesian scan for adaptive genetic divergence in West African cattle. BMC Genomics 10, 550 (2009).
Gautier, M., Laloe, D. & Moazami-Goudarzi, K. Insights into the genetic history of French cattle from dense SNP data on 47 worldwide breeds. PLoS ONE 5, e13038 (2010).
Mi, H. et al. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res 38, D204–D210 (2010).
Strillacci, M. G. et al. Genome-wide association study for somatic cell score in Valdostana Red Pied cattle breed using pooled DNA. BMC Genet 15, 106 (2014).
Tishkoff, S. A. et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 39, 31–40 (2007).
Minozzi, G. et al. Genome wide analysis of fertility and production traits in Italian Holstein cattle. PLoS One 8, e80219 (2013).
García-Fernández, M., Gutiérrez-Gil, B., García-Gámez, E., Sánchez, J. P. & Arranz, J. J. Detection of quantitative trait loci affecting the milk fatty acid profile on sheep chromosome 22: Role of the stearoyl-CoA desaturase gene in Spanish Churra sheep. J Dairy Sci 93, 348–357 (2010).
Chain, F. J. et al. Extensive copy-number variation of young genes across stickleback populations. PLoS Genet 10, e1004830 (2014).
Decker, J. E. et al. Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics. Proc Natl Acad Sci USA 106, 18644–18649 (2009).
Decker, J. E. et al. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet 10, e1004254 (2014).
Xu, L. et al. Genome wide CNV analysis reveals additional variants associated with milk production traits in Holsteins. BMC Genomics 15, 683 (2014).
De Roos, A. P., Hayes, B. J., Spelman, R. J. & Goddard, M. E. Linkage disequilibrium and persistence of phase in Holstein-Friesian, Jersey and Angus cattle. Genetics 179, 1503–1512 (2008).
Nguyen, D. Q., Webber, C. & Ponting, C. P. Bias of selection on human copy-number variants. PLoS Genet 2, e20 (2006).
Handsaker, R. E. et al. Large multiallelic copy number variations in humans. Nat Genet 47, 296–303 (2015).
Falush, D., Stephens, M. & Pritchard, J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 (2007).
Stevens, E. L. et al. Inference of relationships in population data using identity-by-descent and identity-by-state. PLoS Genet 7, e1002287 (2011).
Acknowledgements
We thank members of the Bovine HapMap Consortium for sharing their data. We thank Reuben Anderson and Alexandre Dimitriv for technical assistance. This work was supported in part by AFRI grant No. 2011-67015-30183 from USDA NIFA (G.E.L.) and Youth Innovation Promotion Association, Chinese Academy of Sciences (Y.H.). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. The USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Contributions
G.E.L. and L.X. conceived and designed the experiments. L.X., Y.H., Y.Z., E.H., D.M.B., J.S. and G.E.L. performed in silico prediction and computational analyses. T.S.S. and C.P.V.T. collected samples and generated the S.N.P. genotyping data. G.E.L., L.X. and D.M.B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
Yes there is potential competing financial interests. T.S.S. is an employee of Recombinetics, Inc. All other authors declare no potential conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Xu, L., Hou, Y., Bickhart, D. et al. Population-genetic properties of differentiated copy number variations in cattle. Sci Rep 6, 23161 (2016). https://doi.org/10.1038/srep23161
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep23161
This article is cited by
-
Mapping and functional characterization of structural variation in 1060 pig genomes
Genome Biology (2024)
-
Whole genome resequencing of the Iranian native dogs and wolves to unravel variome during dog domestication
BMC Genomics (2020)
-
Identification and population genetic analyses of copy number variations in six domestic goat breeds and Bezoar ibexes using next-generation sequencing
BMC Genomics (2020)
-
Comparative analyses of copy number variations between Bos taurus and Bos indicus
BMC Genomics (2020)
-
Functional and population genetic features of copy number variations in two dairy cattle populations
BMC Genomics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.