Abstract
Organic acids are essential to fruit flavor. The vacuolar H+ transporting adenosine triphosphatase (V-ATPase) plays an important role in organic acid transport and accumulation. However, less is known of V-ATPase interacting proteins and their relationship with organic acid accumulation. The relationship between V-ATPase and citric acid was investigated, using the citrus tangerine varieties ‘Ordinary Ponkan (OPK)’ and an early maturing mutant ‘Zaoshu Ponkan (ZPK)’. Five V-ATPase genes (CitVHA) were predicted as important to citric acid accumulation. Among the genes, CitVHA-c4 was observed, using a yeast two-hybrid screen, to interact at the protein level with an ethylene response factor, CitERF13. This was verified using bimolecular fluorescence complementation assays. A similar interaction was also observed between Arabidopsis AtERF017 (a CitERF13 homolog) and AtVHA-c4 (a CitVHA-c4 homolog). A synergistic effect on citric acid levels was observed between V-ATPase proteins and interacting ERFs when analyzed using transient over-expression in tobacco and Arabidopsis mutants. Furthermore, the transcript abundance of CitERF13 was concomitant with CitVHA-c4. CitERF13 or AtERF017 over-expression leads to significant citric acid accumulation. This accumulation was abolished in an AtVHA-c4 mutant background. ERF-VHA interactions appear to be involved in citric acid accumulation, which was observed in both citrus and Arabidopsis.
Similar content being viewed by others
Introduction
Organic acids play a pivotal role in plant growth and development; in Arabidopsis, organic acid was observed to be involved in germinating seedlings1. In addition, they participate in a multiplicity of pathways in plant metabolism and are related to metal-resistance2. In fruit, organic acids contribute to flavor, one of the most important fruit quality traits, thus affecting consumer preference. Organic acids also play a crucial role in fruit nutrition and fruit beverage quality3. Concentrations of organic acids are affected by genetic, developmental and environmental stimuli. Among different crops, acidity varies within species or cultivars4. During fruit development, organic acids usually accumulate at early stages and then decrease towards fruit maturity5. Environmental stimuli, such as light and temperature, also influence fruit organic acid metabolism6.
Organic acids, including citric, malic, tartaric, succinic and oxalic acids, are present in various fruits, providing characteristic flavors. In citrus fruits, the predominant organic acid is citric acid. The accumulation of organic acids in fruit is determined by the balance of biosynthesis, degradation and vacuolar storage7. Most research has focused on fruit citric acid biosynthesis and degradation. For citric acid biosynthesis, the activity of mitochondrial citrate synthase (mtCS) was observed to be positively correlated with citrate accumulation in citrus8. However, some reports suggest that the activity and expression of mtCS is not responsible for differences in citrate content between low- and high-acid cultivars9,10. The degradation pathway of citric acid has also been investigated, suggesting that cytosolic aconitase (ACO) and NAD-isocitrate dehydrogenase (NAD-IDH) activities control citric acid degradation during lemon fruit (Citrus limettioides Tan., low acid; Citrus limon var. Eureka, high acid) development11,12. In addition, ACO and IDH, CsGAD1/2 and CitGAD4 are involved in citric acid utilization in orange and mandarin fruit (Citrus sinensis cv. Anliu; C. sinensis cv. Niuher; C. unshiu cv. Guoqing No.113) and hot air driven citric acid degradation in ‘Ponkan’ fruit (C. reticulata Blanco cv. Ponkan14). Following citric acid biosynthesis and degradation, vacuolar storage is important as the flavor of citrus fruit is determined by the concentration of citric acid in vacuole.
Vacuolar accumulation of citric acid is thought to be controlled mainly by vacuolar H+-ATPases15. The vacuolar H+ transporting adenosine triphosphatase (V-ATPase), also known as the ‘proton pump’, is a multi-subunit enzyme using the energy released from the hydrolysis of ATP to pump protons into the vacuolar lumen, thereby creating an electrochemical H+ gradient which is the driving force for a variety of transport events including ions and metabolites, such as organic acids16. The expression of V-ATPase subunit A is associated with loquat fruit acidity, exhibiting higher expression in low-acid loquat fruit ‘Changhong 3’17. Similar results were observed in some other fruit, such as grapes, peaches and apples18,19,20. In citrus, investigation of the vacuoles of the acid lime juice cell suggests citrate uptake can be accounted for by direct primary active transport mechanisms involving ATP21. In addition, the studies in acidic lemon and limes indicated that citric acid accumulation was accompanied by a large influx of protons mediated by the vacuolar H+-ATPase. This influx of protons reduces the vacuolar pH and provides a driving force for additional citric acid uptake15,22. Thus, there is evidence that V-ATPase contribute to citric acid content in citrus fruit. However, the mechanisms were relatively unknown and regulation of the V-ATPase protein complex is not well understood.
In this current study, a novel ethylene response factor (ERF), CitERF13, was shown by yeast two-hybrid assay to interact with the citrus vacuolar proton pump (CitVHA-c4) at the protein-protein level. The correlation of gene expression of CitERF13 and CitVHA-c4 (as well as other coding genes for V-ATPase), with citric acid content was analyzed in two cultivars of Ponkan, a sweet variety of tangerine. The functional characterization of CitERF13 and CitVHA-c4 was analyzed by transient overexpression in tobacco (N. tabacum) and mutants of Arabidopsis. The results suggest that CitERF13 is involved in citric acid accumulation, via protein-protein interaction with CitVHA-c4.
Results
Changes in Organic Acid content during Fruit Development
Organic acids, including citric, tartaric, malic and quinic acid, were measured during Ponkan fruit development (Fig. 1). The results indicate that citric acid is the major component of ‘Ponkan’ fruit organic acids, with 11.90 mg g−1 at maturity in OPK fruit (180 DAFB) (Fig. 1c). Tartaric acid, malic acid and quinic acid contents were lower at the same stage in OPK fruit, with 0.79 mg g−1, 1.12 mg g−1 and 2.97 mg g−1, respectively (Fig. 1d–f).
Changes in the contents of titratable acid (TA,
(a)), total organic acids (b), citric acid (c), tartaric acid (d), malic acid (e) and quinic acid (f) in flesh of Pokan fruits during fruit development, DAFB means day after full blossom. Error bars on each column indicate ±SE from three biological replicates. LSDs represent least significant differences at 0.05.
As citric acid is the major organic acid in ‘Ponkan’ fruits, changes in citric acid content were highly correlated with TA, during fruit development. In both of ZPK and OPK fruit, TA value, total organic acids and citric acid content increased during fruit early developmental stages (peaked at 120 DAFB in OPK fruit) and decreased afterwards till commercial maturity stage (180 DAFB) (Fig. 1a–c). In contrast, the tartaric acid, malic acid and quinic acid contents remained from 60 DAFB to 180 DAFB, with slight changes during development (Fig. 1d–f).
When comparing the two cultivars, the dynamics of organic acid and TA changes were similar between OPK and ZPK. The main difference between OPK and ZPK, was the value of TA and content of total organic acid and citric acid. As shown in Fig. 1, OPK had higher TA, total organic acid and citric acid which peaked with 5.25%, 43.50 mg g−1 and 38.47 mg g−1 at 120 DAFB; while ZPK peaked earlier at 90 DAFB with value of 4.41%, 36.37 mg g−1 and 29.39 mg g−1, respectively. After peak period, TA, total organic acid and citric acid decreased in both cultivar s and the differences between two cultivars were constant till maturity, with value of −1.25%, 15.66 mg g−1 and 11.9 mg g−1 in OPK and was lower in ZPK of −0.82%, 10.20 mg g−1 and 6.74 mg g−1 (Fig. 1a–c)
Expression of V-ATPase Genes during Fruit Development
V-ATPases have been associated with organic acid accumulation. However, the role of V-ATPases in citric acid content in citrus fruit is limited. According to the previous nomenclature (Sze et al., 200223), 18 V-ATPase genes were identified by blast using Arabidopsis V-ATPase genes in the citrus genome database (http://www.citrusgenomedb.org). The 18 V-ATPase genes were designed as CitVHA-A1 (clementine0.9_005081m), CitVHA-A2 (clementine0.9_007969m), CitVHA-B (clementine0.9_008666m), CitVHA-C (clementine0.9_013205m), CitVHA-D (clementine0.9_019158m), CitVHA-E1 (clementine0.9_020736m), CitVHA-F1 (clementine0.9_024363m), CitVHA-F2 (clementine0.9_025353m), CitVHA-G1 (clementine0.9_026050m), CitVHA-H (clementine0.9_030560m), CitVHA-a1 (clementine0.9_002488m), CitVHA-c1 (clementine0.9_023951m), CitVHA-c2 (clementine0.9_023956m), CitVHA-c3 (clementine0.9_023909m), CitVHA-c4 (clementine0.9_023943m, previously known as CitVATP-c1, AB024274.1), CitVHA-c” (clementine0.9_023193m), CitVHA-d (clementine0.9_014414m), CitVHA-e (clementine0.9_027237m). Gene expression analysis indicated that CitVHA genes differentially expressed during fruit development (Fig. 2).
Using OPK as a reference, eight CitVHA genes exhibited increasing expression pattern during fruit development; CitVHA-A1, CitVHA-A2, CitVHA-C, CitVHA-E1, CitVHA-H, CitVHA-a1, CitVHA-c3 and CitVHA-e. Four genes, CitVHA-A2, CitVHA-F1, CitVHA-F2 and CitVHA-G1, showed a peak in expression at 120 DAFB; while some members showed more constitutive expression during development, such as CitVHA-D and CitVHA-d (Fig. 2).
In parallel with citric acid content, differential expression of some CitVHA genes were also observed from two cultivars, such as CitVHA-F1, CitVHA-F2, CitVHA-G1 and CitVHA-a1 were relatively higher expressed at early development stages (60–120 DAFB) in OPK fruit than in ZPK fruit (Fig. 2). In contrast, several CitVHA genes, such as CitVHA-E1, CitVHA-c3, CitVHA-c” and CitVHA-e, were highly expressed in ZPK fruit at late development stage (Fig. 2). Of interest was CitVHA-c4, which showed substantially higher expression in OPK fruit than in ZPK fruit (Fig. 2). Thus, CitVHA-c4, could be a candidate responsible for differences in citric acid content in ‘Ponkan’ fruit.
A transcription factor, CitERF13, interacts with CitVHA-c4
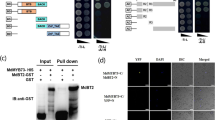
A transcriptome-wide screening experiment was performed to identify potential proteins that could interact with the closest Arabidopsis homologue to CitERF13, AtERF017 (At1g19210), using the commercial yeast two-hybrid library from DUALhunter systems Biotech (Switzerland). A protein was identified which shown to be AtVHA-c4 (At1g75630), that interacted with AtERF017 (Fig. 3). Phylogenetic analysis indicated that AtVHA-c4 is a homolog of CitVHA-c4 (Fig. S2). Thus, it is proposed that ERF transcription factor may act novel regulator of citric acid metabolism via binding to the V-ATPase.
Yeast two-hybrid assays show that AtERF017 interacts with AtVHA-c4 and CitERF13 interacts with CitVHA-c4.
AtERF017 and AtVHA-c4 are homologs of CitERF13 and CitVHA-c4 in Arabidopsis. Liquid cultures of double transformants are plated at OD600 = 0.1 dilutions of the cultures on synthetic dropout selective medium: (1) SD medium lacks Trp and Leu (DDO); (2) SD medium lacks Trp, Leu, His and Ade (QDO); (3) SD medium lacks Trp, Leu, His, Ade and was supplemented with 10 mM 3-amino-1,2,4-triazole (QDO+3AT). Protein-protein interactions were determined on QDO and QDO + 3AT. pOst1-NubI, positive control; pPR3-N, negative control.
Within the CitAP2/ERF family, an ethylene response factor (CitERF13, isolated by Xie et al.24 was found as the closest homolog of AtERF017, based on phylogenetic analysis (Fig. S2). Thus, potential protein-protein interaction between CitERF13 and CitVHA-c4 was analyzed. Similar to AtERF017, CitERF13 could also interact with CitVHA-c4 (Fig. 3). Moreover, further analyses indicated that such interaction only occurred between CitERF13 and CitVHA-c4, but not the others c units, including CitVHA-c1, CitVHAc2, CitVHA-c3 and CitVHA-c” (Fig. S3). This result, suggests that CitERF13 and AtERF017 participate in citric acid regulation via a specific interaction with one subunit of the V-ATPase.
To further confirm the interaction between CitERF13 and CitVHA-c4 and to verify the in vivo interactions in planta, bimolecular fluorescence complementation assays were performed. The results showed that the negative combination, such as CitERF13-YFPN/YFPC, YFPN/CitERF13-YFPC, CitVHA-c4-YFPN/YFPC and YFPN/CitVHA-c4-YFPC and YFPN/YFPC did not produce any detectable fluorescence signal, while co-expression of CitERF13-YFPN and CitVHA-c4-YFPC or CitVHA-c4-YFPN and CitERF13-YFPC gave strong signals both in the nucleus and at the tonoplast, such fluorescence detection indicated protein-protein interaction between CitERF13 and CitVHA-c4 and further supported yeast two hybrid results (Fig. 4).
In vivo interaction between CitERF13 and CitVHA-c4, using BiFC.
BiFC analysis for interaction between CitERF13 and CitVHA-c4. N- and C-terminal fragments of YFP (YFPN and YFPC) were fused to the C terminus of CitERF13 and CitVHA-c4, respectively. Combination of YFPC or YFPN with corresponding CitERF13 or CitVHA-c4 constructs were used as negative controls. Fluorescence of YFP represents protein-protein interaction. The bars indicated the length of 50 μm.
Association of CitERF13 expression and citric acid during fruit development
In order to confirm the relationship between CitERF13 and citric acid, the expression of CitERF13 was studied during fruit development in OPK and ZPK. The results indicated that CitERF13 had a similar expression pattern to CitVHA-c4, which was more abundant in the high-acidity OPK fruit during fruit development (Fig. 5). Furthermore, other genetically unrelated citrus material was checked. It was found that CitERF13 exhibited higher expression in the high-acidity ‘Gaocheng’ Mandarin than in low-acidity ‘Satsuma Mandarin’25 (Fig. S4).
Effects of CitERF13 and CitVHA-c4 on in citric acid accumulation
As citrus is a perennial fruit, thus transient overexpression analyses were conducted N. tabacum leaves to verify CitERF13 and CitVHA-c4 function26. Compared with empty vector control, transient overexpression of CitERF13 and CitVHA-c4 significantly increased the citric acid accumulation in tobacco leaves. Citric acid in N. tabacum, transformed with Agrobacterium harboring the empty vector alone, was not detectable. With transformation of the leaves with either CitERF13 or CitVHA-c4 alone, the citric acid in N. tabacum leaves reached 0.52 mg g−1 and 0.25 mg g−1, respectively (Fig. 6). Moreover, a combination of CitERF13 and CitVHA-c4 accumulated more citric acid, reaching 0.68 mg g−1 (Fig. 6).
Transient overexpression of CitERF13 and CitVHA-c4 in N. tabacum leaves.
Both CitERF13 and CitVHA-c4 genes were driven by the CaMV 35S promoter. SK represents empty vector. Citric acid was analyzed at 5 d after infiltration, with three biological replicates. Different letters indicate a significant difference (p < 0.05), which were calculated using Student’s t-test. +, present of the construct; −, absence of the construct.
The function of the ERF and its interaction with the V-ATPase, were further confirmed in Arabidopsis AtVHA-c4 mutant. An AtVHA-c4 mutant (CS548548) was obtained from TAIR and real-time PCR analysis indicated the endogenous AtVHA-c4 had relatively low expression in the mutant (CS848548), with approximately 75% reduction of that in WT plants (Fig. 7c). Transient overexpression of AtERF017 significantly increased citric acid accumulation; in wild type Col Arabidopsis leaves, with transient expression of AtERF017, citric acid reaches 0.18 mg g−1, compared with only 0.11 mg g−1 in the leaves infiltrated with empty vector. However, in the AtVHA-c4 mutant CS848548, AtERF017 transient overexpression failed to enhance citric acid accumulation, compared with empty vector (Fig. 7a). Similar results were found using CitERF13, the homolog of AtERF017, which also exhibited significant enhancement of citric acid level in Arabidopsis leaves (Fig. 7b).
Transient overexpression of AtERF017 and CitERF13 in Arabidopsis leaves.
CS848548 is a mutant for AtVHA-c4 and was obtained from TAIR. (a,b) Citric acid was analyzed at 3 d after infiltration, with three biological replicates. The statistical significance of differences was calculated using Student’s t-test. Asterisks *indicate significant differences (p < 0.05), double asterisks **indicate significant differences (p < 0.01). (c) AtVHA-c4 mRNA abundance in wild type and CS848548. AtERF017 and CitERF13 gene was driven by the CaMV 35S promoter. SK represents empty vector.
Discussion
Most organic acids are found in the plant vacuole, which is considered as the main reservoir of these metabolites27. As one of the most important enzymes, the V-ATPase, therefore plays a very important role in organic acid accumulation. The V-ATPase is a large multimeric enzyme organized in two domains, V1 and V0. The peripheral V1 domain is responsible for hydrolysis of ATP and was comprised of eight subunits (A-H), while V0 domain is composed of six different subunits a, d, c, c’ (absent in plants), c”, e, which function on translocation of protons across the membrane28,29. Each subunit has dominant functions for some specific processes, eg. VHA-E1 plays an essential role in maintaining a functional secretory system during somatic development30; CsVHA-c1 and CsVHA-c2 were essential elements of mechanisms involved in adaptation of cucumber plants to copper toxicity31; MdVHA-A and MdVHA-B from apple were observed to be involved in drought tolerance32,33. However, most research has focused on transcriptional response of VHA genes in response to environmental stimuli and the functional characterization of each subunit. The linkage between V-ATPases and transcription factors has not been investigated.
Here, 18 genes encoding subunits of V-ATPase were isolated from the citrus genome database. In order to relate gene expression with organic acid levels, OPK and ZPK were chosen as material, as ZPK was considered as a mutant bud sport of OPK, thus ZPK and OPK should had similar genetic backgound. It is interesting that ZPK and OPK had a very similar profile of organic acid change, but ZPK had lower organic acid (mainly citric acid) than OPK fruit. Gene expression suggested indicated that CitVHA-F1, CitVHA-F2, CitVHA-G1, CitVHA-a1 and CitVHA-c4 were associated with the citric acid content in OPK and ZPK. Similar results were reported in ‘sweet’ and ‘sour’ lemon, in which expression of the H+-ATPase AHA10 is higher in sour lemon and may be involved in citric acid biosynthesis and accumulation in juice sac cells34. These results provided further molecular evidence for the role of V-ATPase in citric acid and those five genes (CitVHA-F1, CitVHA-F2, CitVHA-G1, CitVHA-a1 and CitVHA-c4 might be the key dominant unit for different acidity in citrus fruit.
CitVHA-c4 was constitutively higher in expression in OPK fruit, which correlates with higher citric acid content. A similar correlation of CitVHA-c4 expression was also observed using high-acid ‘Gaocheng’ (GC) and the low-acid ‘Satsuma Mandarin’ (SM) (Fig. S4). Transient over-expression of CitVHA-c4 in N. tabacum leaves significantly elevated citric acid accumulation. Thus, it is proposed that CitVHA-c4 is an important unit for citric acid accumulation, which contributes to citrus fruit acidity and flavor.
One of most interesting results from this study is the novel protein-protein interaction observed between CitERF13 and CitVHA-c4. Such an interaction was further confirmed in Arabidopsis between AtERF017 (a homolog of CitERF13) and AtVHA-c4 (a homolog of CitVHA-c4), suggesting that the interaction between AP2/ERF and V-ATPase might be conserved within plants. AP2/ERFs are a large plant specific transcription factor family, which are involved in many aspects of plant development and stress response35,36. In fruit, various AP2/ERF genes were characterized to be involved in fruit ripening37,38 and quality aspects (eg. Carotenoid39; texture40; taste41,42). No association of an ERF and V-ATPase has been reported. However, some other well-known proteins function via the V-ATPase, such as the protein kinase SOS2 which promotes salt tolerance by interacting with VHA-B subunits43. The regulatory 14-3-3 proteins are activators which interact with the A-subunit of the V-ATPase after blue light treatment44. In addition, different subunits could also interact with each other, such as subunit c and a45. Therefore, citrus CitERF13 and Arabidopsis AtERF017 appear to have a novel interaction with the V-ATPase.
Using GFP tags CitERF13 and CitVHA-c4 were predicted to have different subcellular localizations. It has been reported in Arabidopsis that the ethylene-responsive element binding protein (AtEBP) may move from the nucleus to the cytosol via protein-protein interaction with ACBP446; a rice SPX family protein, OsSPX4, can also reduce the targeting of OsPHR2 to the nucleus through its interaction with PHR2 when Pi is sufficient47. In our citrus study, a protein-protein interaction between CitERF13 and CitVHA-c4 was verified using BiFC assays. This indicated a strong signal in both the tonoplast and nucleus. Thus, interaction between CitERF13 and CitVHA-c4 may shift their sub-cellular localization. Importantly, CitERF13 and its homolog AtERF017, both exhibited the ability to enhance citric acid accumulation. Transient over-expression of CitERF13 in tobacco triggered citric acid accumulation. Moreover, combinatorial effects of CitERF13 and CitVHA-c4, as well as AtERF017 and AtVHA-c4, were observed. A combination of CitERF13 and CitVHA-c4 infiltrated into tobacco resulted in an increment (with significance), compared with CitERF13 and CitVHA-c4 infiltrated individually. These results suggested that CitERF13 and CitVHA-c4 work as partners in citric acid accumulation. Both of AtERF017 and CitERF13 was failed to trigger citric acid accumulation in leaves of CS838548, where AtVHA-c4 was significantly repressed. Taken together, these results suggest CitERF13, as well as Arabidopsis AtERF017, are novel regulators on citric acid accumulation, via the V-ATPase c unit.
In conclusion, our results indicated that in citrus, CitVHA-F1, CitVHA-F2, CitVHA-G1, CitVHA-a1 and CitVHA-c4 are related to the citric acid accumulation. A novel protein-protein interaction was observed between CitERF13 and CitVHA-c4, with such a complex linking CitERF13 and citric acid accumulation. Although such protein-protein interactions are rarely reported in plants, the present results showed similar interactions between AtERF017 and AtVHA-c4. Thus this protein-protein interaction might be conserved for various plants.
Methods
Plant materials
Fruit of two cultivars of Ponkan (C. reticulata Blanco cv. Ponkan), named as ‘Ordinary Ponkan (OPK)’ and ‘Zaoshu Ponkan (ZPK)’, were harvested from a commercial orchard in Quzhou, Zhejiang, China. ZPK is a bud sport mutant of OPK, which in this region is early maturing. Fruits with uniform size and appearance were collected for each cultivar, at each sampling point, from six different trees. Six time points were collected at 60, 90, 120, 150, 165 and 180 days after full bloom (DAFB). The flesh was frozen in liquid nitrogen and stored at −80 °C for further experiments.
Titratable acidity and organic acid measurement
Fruits were divided into three groups, each of four fruits and 5 ml of juice from each group was diluted in 20 ml distilled water and then was titrated with 0.1 M NaOH to the end point at pH 8.2, according to the previous report48. Titratable Acidity (TA) was calculated as percentage of citric acid.
Organic acids were extracted according to the method described by Chen et al.14. Two grams of frozen flesh sample was ground to a powder in liquid nitrogen and homogenized in 5.0 ml of ethanol (80%) at 35 °C for 20 min. The homogenate was centrifuged at 10,000 × g for 10 min, at 20 °C. The residue was extracted twice and the supernatant was collected and supplemented with 80% ethanol to 25 ml. One milliliter extracted solution was dried under vacuum condition (Eppendorf Concentrate Plus, Germany) at 45 °C and the residue was dissolved in 0.5 ml distilled water and filtered with Ф0.22 μm, ø13 mm water syringe filter (Shanghai Xingya Purification Material Factory, China). The filtered solution was used for further organic acids analysis.
Organic acids were analyzed by high-performance liquid chromatography (HPLC) (Waters Alliance 2695 system, Waters Corporation, USA). A 20 μl sample of eluate was injected into an ODS C18 (4.6 × 250 mm) column (Beckman, USA). The flow rate was 0.5 ml min−1 using 50 mM (NH4)2HPO4 (pH 2.7, which was adjusted by H3PO4) as the solvent. Organic acids were detected at 210 nm. The eluted peaks were detected with a Waters 2996 diode array detector (Waters Corporation, USA) and quantity of individual organic acids was calculated using peak areas of standards. All measurements for organic acids were performed with three replicates.
RNA extraction and cDNA synthesis
Total RNA was extracted from frozen tissues according to the protocol described by Chang49. The genomic DNA in total RNA was degraded with RNase-free DNase I (Fermentas). A 1.0 μg DNA-free RNA was initiated for first-strand cDNA synthesis with a RevertAid™ Premium Reverse Transcriptase (Fermentas, Thermo Scientific, USA) following to the manufacturer’s protocol. Diluted cDNA was used as the template for real-time PCR analysis. RNA extraction and cDNA synthesis were performed with three biological replicates for each sampling point.
Real-time PCR
The PCR mixture (10 μL total volume) comprised 2 μL of Lightcycler Faststart DNA Masterplus SYBR Green I Mix (Roche), 0.5 μL of each primer (10 mM), 1 μL of diluted cDNA and 6 μL PCR grade H2O. PCR was performed on a LightCycler 1.5 instrument (Roche), initiated by 5 min at 95 °C, then followed by 45 cycles of 95 °C for 10 s, 60 °C for 5 s and 72 °C for 10 s and completed with a melting curve analysis program. No-template controls and melting curve analyses were included in every reaction. Citrus actin (XM_006464503) was used as the housekeeping gene to quantify cDNA abundance14. The sequences of primers for CitVHA, AtERF017 and AtVHA-c4 were described in Table S1.
Yeast two-hybrid assay
Protein-protein interactions were investigated in yeast with the DUAL hunter system (Dualsystems Biotech, Switzerland). Full-length coding sequences of AtERF017 and CitERF13 were cloned into pDHB1 vector, as bait; while full-length of AtVHA-c4 and CitVHA-c subunits were constructed into pPR3N vector, as prey. The primers used for vectors construction were described in Table S2.
All constructs were transformed in the yeast strain NMY51 according to the manufacturer’s instructions. The assays were performed with different mediums: (1) SD medium lacks Trp and Leu (DDO); (2) SD medium lacks Trp, Leu, His and Ade (QDO); (3) SD medium lacks Trp, Leu, His, Ade and was supplemented with 10 mM 3-amino-1,2,4-triazole (QDO + 3AT). Auto-activations were tested with empty vector of pPR3-N and target genes with pDHB1, which were co-transformed in NMY51 and plated on QDO. Auto-activations were indicated by presence of colonies. Protein-protein interaction assays were performed with co-transformation of AtVHA-c4 in pPR3N with AtERF017 in pDHB1 and CitVHA-c subnuits in pPR3N with CitERF13 in pDHB1, respectively. The presence of colonies in QDO and QDO + 3AT, indicated a protein-protein interaction.
Bimolecular fluorescence complementation assay
Full-length CitERF13 and full-length CitVHA-c4 were cloned into either C-terminal or N-terminal fragments of YFP vectors50. Primers used are listed in Supplemental Table 2. All constructs were transiently expressed in tobacco leaves by Agrobacterium-mediated infiltration (GV3101) according to previous reports51. The YFP fluorescence of tobacco leaves were imaged 3 d after infiltration using a Zeiss LSM710NLO confocal laser scanning microscope. The excitation wavelength for YFP fluorescence was 514 nm and fluorescence was detected at 519 to 567 nm.
Transient over-expression in tobacco and Arabidopsis
Full-length coding sequences of CitERF13 and CitVHA-c4 were amplified with primers (listed in Supplementary Table 3) and were constructed into pGreen II 0029 62-SK vector (SK). The detail information of the SK vector was described in Hellens et al.27. The constructs were electroporated into Agrobacterium GV3101. Agrobacterium cultures, carrying empty vector (SK), CitERF13, CitVHA-c4 and CitERF13 + CitVHA-c4, were infiltrated into different sites of same leaf, as indicated in Figure S1. Five days after infiltration, the infiltrated leaves were sampled and used for citric acid analysis.
Furthermore, transient over-expression was also conducted in Arabidopsis leaves, to verify the function of AtERF017, a homolog of CitERF13. Seeds of AtVHA-c4 mutant (CS838548) were obtained from The Arabidopsis Information Resource (TAIR). Full-length of AtERF017 was obtained with primers listed in Table S3 and was also constructed in the SK vector. Using the same protocol of tobacco system, Agrobacterium, carrying AtERF017 and empty vector (SK), were transiently over-expressed in leaves of wild type (Col) and the two mutant lines. Infiltrated leaves were sampled and used for citric acid analysis.
The citric acid content of infiltrated leaves of N. tabacum and Arabidopsis were measured according Lin et al.52. Leaves (0.05 g) were ground in liquid nitrogen and extracted with 1.4 ml of methanol, at 70 °C for 15 min and then centrifuged at 10,000 g. The upper phase was removed and stored at −80 °C until analysis. Aliquots of 100 μl upper phase were dried in vacuum. The residue was dissolved in 40 μl 20 mg ml−1 pyridine methoxyamine hydrochloride and incubated at 37 °C for 1.5 h. The sample was then treated with 60 μl Bis (trimethylsilyl) trifluoroacetamide (1% trimethylchlorosilane) at 37 °C for 30 min. Ribitol (20 μl, 0.2 mg ml−1) was added into each sample as an internal standard. A 1 μl aliquot of each sample was absorbed with a split ratio of 1:1 and injected into GC-MS fitted with a fused-silica capillary column (30 m × 0.25 mm i.d., 0.25 μm DB-5 MS stationary phase). The injector temperature was 250 °C and the helium carrier gas had a flow rate of 1.0 ml min−1. The column temperature was held at 100 °C for 1 min, increased to 184 °C with a rate of 3 °C min−1, then increased to 230 °C with rate of 15 °C min−1, held for 1 min. The MS operating parameters were as follows: ionization voltage was 70 eV, ion source temperature was 230 °C and the interface temperature was 280 °C.
Statistical Analysis
Least significant difference (LSD) was calculated by DPS 7.05 (Zhejiang University, Hangzhou, China). The statistical significance of differences was calculated using Student’s t-test.
Additional Information
How to cite this article: Li, S.-j. et al. The Citrus transcription factor, CitERF13, regulates citric acid accumulation via a protein-protein interaction with the vacuolar proton pump, CitVHA-c4. Sci. Rep. 6, 20151; doi: 10.1038/srep20151 (2016).
References
Hooks, M. A., Turner, J. E., Murphy, E. C. & Graham, I. A. Acetate non-utilizing mutants of Arabidopsis: evidence that organic acids influence carbohydrate perception in germinating seedlings. Mol. Gen. Genomics 271, 249–256 (2004).
Ding, Z. J., Yan, J. Y., Xu, X. Y., Li, G. X. & Zheng, S. J. WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 76, 825–835 (2013).
Zampini, M., Wantling, E., Phillips, M. & Spence, C. Multisensory flavor perception: Assessing the influence of fruit acids and color cues on the perception of fruit-flavored beverages. Food Qual. Prefer. 19, 335–343 (2008).
Scherer, R. et al. Validation of a HPLC method for simultaneous determination of main organic acids in fruits and juices. Food Chem. 135, 150–154 (2012).
Chen, F. X., Liu, X. H. & Chen, L. S. Developmental changes in pulp organic acidconcentration and activities of acid-metabolising enzymes during the fruit development of two loquat (Eriobotrya japonica Lindl.) cultivars differing in fruit acidity. Food Chem. 114, 657–664 (2009).
Biais, B. et al. Remarkable reproducibility of enzyme activity profiles in tomato fruits grown under contrasting environments provides a roadmap for studies of fruit metabolism. Plant Physiol. 164, 1204–1221 (2014).
Wang, X. Y. et al. Effects of granulation on organic acid metabolism and its relation to mineral elements in Citrus grandis juice sacs. Food Chem. 145, 984–990 (2014).
Sadka, A. et al. Comparative analysis of mitochondrial citrate synthase gene structure, transcript level and enzymatic activity in acidless and acid-containing citrus varieties. Aus. J. Plant Physiol. 28, 383–390 (2001).
Canel, C., BaileySerres, J. N. & Roose, M. L. Molecular characterization of the mitochondrial citrate synthase gene of an acidless pummelo (Citrus maxima). Plant Mol. Biol. 31, 143–147 (1996).
Yu, K. Q. et al. Transcriptome changes during fruit development and ripening of sweet orange (Citrus sinensis). BMC Genomics 13, 10 (2012).
Sadka, A., Dahan, E., Cohen, L. & Marsh, K. B. Aconitase activity and expression during the development of lemon fruit. Physiol. Plantarum 108, 255–262 (2000).
Sadka, A., Dahan, E., Or, E. & Cohen, L. NADP+-isocitrate dehydrogenase gene expression and isozyme activity during citrus fruit development. Plant Sci. 158, 173–181 (2000).
Liu, X. et al. Identification and transcript analysis of two glutamate decarboxylase genes, CsGAD1 and CsGAD2, reveal the strong relationship between CsGAD1 and citrate utilization in citrus fruit. Mol. Biol. Rep. 41, 6253–6262 (2014).
Chen, M. et al. Effect of hot air treatment on organic acid- and sugar metabolism in Ponkan (Citrus reticulata) fruits. Sci. Hortic. 147, 118–125 (2012).
Müller, M. L., IrkensKiesecker, U., Kramer, D. & Taiz, L. Purification and reconstitution of the vacuolar H+-ATPases from lemon fruits and epicotyls. J. Membrane Biol. 272, 12762–12770 (1997).
Ratajczak, R. Structure, function and regulation of the plant vacuolar H+-translocating ATPase. BBA-Biomembranes 1465, 17–36 (2000).
Yang, L. T., Xie, C. Y., Jiang, H. X. & Chen, L. S. Expression of six malate-related genes in pulp during the fruit development of two loquat (Eriobotrya japonica) cultivars differing in fruit acidity. Afr. J. Biotechnol. 10, 2414–2422 (2011).
Terrier, N., Sauvage, F. X., Ageorages, A. & Romieu, C. Changes in acidity and in proton transport at the tonoplast of grape berries during development. Planta 213, 20–28 (2001).
Etienne, C. et al. Isolation and characterization of six peach cDNAs encoding key proteins in organic acid metabolism and solute accumulation: involvement in regulating peach fruit acidity. Physiol. Plantarum 114, 259–270 (2002).
Yao, Y. X. et al. Molecular cloning of three malic acid related genes MdPEPC, MdVHA-A, MdcyME and their expression analysis in apple fruits. Sci. Horti. 122, 404–408 (2009).
Brune, A., Gonzalez, P., Goren, R., Zehavi, U. & Echeverria, E. Citrate uptake into tonoplast vesicles from acid lime (Citrus aurantifolia) juice cells. J. Membrane Biol. 166, 197–203 (1998).
Müller, M. L. & Taiz, L. Regulation of the lemon-fruit V-ATPase by variable stoichiometry and organic acids. J. Membrane Biol. 185, 209–220 (2002).
Sze, H., Schumacher, K., Muller, M. L., Padmanaban, S. & Taiz, L. A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase. Trends Plant Sci. 7, 157–161 (2002).
Xie, X. L. et al. Isolation, classification and transcription profiles of the AP2/ERF transcription factor superfamily in citrus. Mol. Biol. Rep. 41, 4262–4271 (2013).
Lin, Q. et al. Involvement of CitCHX and CitDIC in developmental-related and postharvest-hot air driven citrate degradation in Citrus Fruits. PLos ONE 10, e0119410 (2015).
Hellens, R. P. et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 132005 (2005).
Etxeberria, Ed., Pozueta-Romero, J. & Gonzalez, P. In and out of the plant storage vacuole. Plant Sci. 190, 52–61 (2012).
Jefferies, K., Cipriano, D. & Forgac, M. Function, structure and regulation of the vacuolar H+-ATPase. Arch. Biochem. Biophys. 476, 33–42 (2008).
Toei, M., Saum, R. & Forgac, M. Regulation and isoform function of the V-ATPases. Biochemistry 49, 4715–4723 (2010).
Strompen, G. et al. Arabidopsis vacuolar H+-ATPase subunit E isoform 1 is required for Golgi organization and vacuole function in embryogenesis. Plant J. 41, 125–132 (2005).
Kabała, K., Janicka-Russak, M., Reda, M. & Migocka, M. Transcriptional regulation of the V-ATPase subunit c and V-PPase isoforms in Cucumis sativus under heavy metal stress. Physiol. Plantarum 150, 32–45 (2014).
Hu, D. G. et al. Overexpression of MdVHA-B, a V-ATPase gene from apple, confers tolerance to drought in transgenic tomato. Sci Hortic 145, 94–101 (2012).
Dong, Q. L. et al. MdVHA-A encodes an apple subunit A of vacuolar H+-ATPase and enhances drought tolerance in transgenic tobacco seedlings. J. Plant Physiol. 170, 601–609 (2013).
Aprile, A. et al. Expression of the H+-ATPase AHA10 proton pump is associated with citric acid accumulation in lemon juice sac cells. Funct. Integr. Genomics 11, 551–563 (2011).
Zhang, L. X. et al. An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 157, 854–865 (2011).
Licausi, F., Ohme-Takagi, M. & Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649 (2013).
Yin, X. R., Allan, A. C., Chen, K. S. & Ferguson, I. B. Kiwifruit E. I. L. and ERF genes involved in regulating fruit ripening. Plant Physiol. 153, 1280–1292 (2010).
Xiao, Y. Y. et al. Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J. Exp. Bot. 64, 2499–2510 (2013).
Lee, J. M. et al. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 70, 191–204 (2012).
Zeng, J. K. et al. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J., in press (2015).
Min, T. et al. Ethylene-responsive transcription factors interact with promoters of ADH and PDC involved in persimmon (Diospyros kaki) fruit de-astringency. J. Exp. Bot. 63, 6393–6405 (2012).
Min, T. et al. Two novel anoxia-induced ethylene response factors that interact with promoters of deastringency-related genes from persimmon. PLos ONE 9, 97043 (2014).
Batelli, G. et al. SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell Biol. 27, 7781–7790 (2007).
Klychnikov, O. I., Li, K. W., Lill, H. & Boer, A. H. The V-ATPase from etiolated barley (Hordeum vulgare L.) shoots is activated by blue light and interacts with 14-3-3 proteins. J. Exp. Bot. 58, 1013–1023 (2007).
Seidel, T. et al. Colocalization and FRET-analysis of subunits c and a of the vacuolar H+-ATPase in living plant cells. J. Biotechnol. 112,165–175 (2004).
Li, H. Y., Xiao, S. & Chye, L. M. Ethylene- and pathogen-inducible Arabidopsis acyl-CoAbinding protein 4 interacts with an ethylene-responsive element binding protein. J. Exp. Bot. 59, 3997–4006 (2008).
Lv, Q. D. et al. SPX4 Negatively Regulates Phosphate Signaling and Homeostasis through Its Interaction with PHR2 in Rice. Plant Cell 26, 1586–1597 (2014).
Cai, C. et al. Effect of 1-MCP on postharvest quality of loquat fruit. Posthaevest Biol. Technol. 40,155–162 (2006).
Chang, S. J., Puryear, J. & Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11,113–116 (1993).
Sainsbury, F., Thuenemann, E. C. & Lomonossoff, G. P. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693 (2009).
Xu, Q. et al. Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 65, 4349–4359 (2014).
Lin, Q. et al. Transcriptome and metabolome analyses of sugar and organic acid metabolism in Ponkan (Citrus reticulata) fruit during fruit maturation. Gene 544, 64–74 (2015).
Acknowledgements
This research was supported by the National Basic Research Program of China (2011CB100602) and Program of International Science and Technology Cooperation (2011DFB31580), the National High Technology Research and Development Program of China (2012AA101702).
Author information
Authors and Affiliations
Contributions
X.Y. and K.C. designed and supervised the study. S.L., X.X., H.G. and S.S. carried out the experiments and data interpretation. X.Y., A.A. and K.C. interpreted the data and wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Li, Sj., Yin, Xr., Xie, Xl. et al. The Citrus transcription factor, CitERF13, regulates citric acid accumulation via a protein-protein interaction with the vacuolar proton pump, CitVHA-c4. Sci Rep 6, 20151 (2016). https://doi.org/10.1038/srep20151
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep20151
This article is cited by
-
Weighted gene coexpression correlation network analysis reveals the potential molecular regulatory mechanism of citrate and anthocyanin accumulation between postharvest ‘Bingtangcheng’ and ‘Tarocco’ blood orange fruit
BMC Plant Biology (2023)
-
Regulation of a vacuolar proton-pumping P-ATPase MdPH5 by MdMYB73 and its role in malate accumulation and vacuolar acidification
aBIOTECH (2023)
-
Identification of key gene networks controlling organic acid and sugar metabolism during watermelon fruit development by integrating metabolic phenotypes and gene expression profiles
Horticulture Research (2020)
-
A low citric acid trait in cranberry: genetics and molecular mapping of a locus impacting fruit acidity
Tree Genetics & Genomes (2020)
-
Transcriptomic analysis of Citrus clementina mandarin fruits maturation reveals a MADS-box transcription factor that might be involved in the regulation of earliness
BMC Plant Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.