Abstract
Preeclampsia (PE) is a leading cause of maternal mortality worldwide. Several studies have detected some differentially expressed microRNAs in the preeclamptic placenta, but few of the identified microRNAs demonstrated consistent findings among different research studies. In this study, high-throughput microRNA sequencing (HTS) of 9 preeclamptic and 9 normal placentas was performed. Seventeen microRNAs were identified to be up-regulated and 8 down-regulated in preeclamptic placentas. Eight differentially expressed microRNAs except one identified in our study were determined to be consistent with at least one previous study, while sixteen were newly found. We performed qRT-PCR with independent 22 preeclamptic placentas and 20 control placentas to verify the differentially expressed microRNAs and ten microRNAs were validated. The predicted target genes of the aberrantly expressed miR-193b-3p were enriched in the following gene ontology categories: cell motility and migration, cell proliferation and angiogenesis. We also found that miR-193b-3p significantly decreased the migration and invasion of trophoblast (HTR-8/SVneo) cells and that miR-193b-3p could regulate trophoblasts migration and invasion through binding onto the 3′UTR target site of TGF-β2. In conclusion, we identified a list of differentially expressed microRNAs in PE placentas by HTS and provided preliminary evidence for the role of miR-193b-3p in the pathogenesis of preeclampsia.
Similar content being viewed by others
Introduction
Preeclampsia (PE) is characterized by the de-novo development of hypertension and proteinuria at ≥20 weeks of gestation1. This pregnancy-specific syndrome complicates approximately 2% to 8% of pregnancies and is a leading cause of maternal mortality worldwide2. Delivery of the placenta is the only effective treatment for preeclampsia, indicating that the placenta is indispensable to the development of preeclampsia3. According to the traditional view of the pathogenetic mechanisms involved in preeclampsia, initial insults disrupt the deep invasion of the trophoblast resulting in shallow implantation and abnormal remodelling of the placental spiral artery long before 12 to 20 weeks of gestation and in response to poor placentation, proinflammatory and antiangiogenic factors from the foetal/placental unit modify maternal physiology manifesting the clinical characteristics of PE4,5,6,7.
MicroRNAs(MiRNAs) are non-coding RNAs with approximately 21–25 nucleotides in length, which are estimated to post-transcriptionally regulate the expression of nearly 30% of all genes in animals by cleavage or translational interference8. MiRNAs are involved in regulating trophoblast proliferation, apoptosis, migration and invasion and have been suggested to play an important role in the regulation of placental development and function9. Several studies based on hybridization, RT-qPCR or sequencing analysis have detected a list of differentially expressed miRNAs in PE placenta. However, only limited number of these identified miRNAs demonstrated consistent findings among these studies. For instance, six miRNA profiling research analyses showed that miR-210 was aberrantly expressed10,11,12,13,14,15 and three showed that miR-193b-3p consistently up-regulated in PE placentas10,11,12. Efforts have been made to identify how differentially expressed miRNAs contribute to the onset of PE. Evidence suggested that dysfunction of miRNAs could inhibit migration and invasion of human extravillous trophoblast-derived HTR-8/SVneo cells16,17. However, it is not yet the right time to use these findings from miRNA studies on preeclampsia to improve the management or early recognition of this condition. Further investigation is required to elucidate novel mechanisms underlying the molecular pathology of PE by specific miRNAs which have not been investigated, like miR-193b-3p.
High-throughput sequencing (HTS) platforms perform well in reproducibility, accuracy, specificity and sensitivity for quantitatively measuring RNA. In addition, the detection sensitivity has been shown to be increased along with increasing sequencing depth, which is superior to hybridization-based platforms, whereas RT-qPCR is limited by the number of microRNAs18. Four studies conducted by Guo et al. 201119, Ishibashi et al. 201212, Weedon-Fekjær et al. 201420 and and Yang et al. 201521 also performed miRNA profiling of PE placentas using sequencing platforms. However, three12,19,21 performed the research based on limited sample sizes and one20 conducted the research with limited sequencing depth.
In the present study, we performed HTS analysis on 9 preeclamptic and 9 normal placentas with enough sequencing depth to explore the differentially expressed miRNAs and further investigate their biological roles in the development of PE. Our study demonstrated that the expression of miR-193b-3p was significantly up-regulated in PE placentas and it significantly decreased the migration and invasion of HTR-8/SVneo cells. Furthermore, we found that transforming growth factor-beta 2 (TGF-β2) was post-transcriptionally dysregulated by miR-193b-3p and confirmed that miR-193b-3p could regulate trophoblast cell invasion and migration by targeting TGF-β2.
Results
Characteristics of microRNA expression by HTS
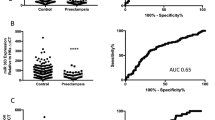
We performed HTS on 9 preeclamptic placentas (P1–P9) and 9 normal placentas (control group, C1–C9). We analyzed miRNAs with ≥5 sequence reads and constructed a scatter plot of these 787 miRNAs with the normalized reads (per million). Comparing global miRNA expression profiles in placentas of preeclamptic patients with those of normotensive controls, we could intuitively observe the distribution of expression of the two groups (Fig. 1A). The majority of miRNAs were not significantly differentially expressed between control and preeclampsia subjects and the expression level of the majority of miRNAs was low (below 500 reads per million reads). Top 30 miRNAs constituted around 80% of all miRNA expression in normal and preeclamptic placenta tissues (Fig. 1B, Supplementary Table S1).
Characteristics of placental microRNA expression profiles in normotensive and preeclamptic pregnancy.
Global placental microRNA expression distribution (sequence reads ≥5) of normotensive controls (n = 9) and preeclamptic patients (n = 9) by HTS (A). The top 30 microRNAs constitute ~80% of all differentially expressed microRNAs between control and preeclamptic placentas (B).
Differentially expressed microRNAs in placentas of patients with preeclampsia
A total of 25 miRNAs were significantly deregulated, 17 of which were up-regulated and 8 were down-regulated in preeclampsia (Table 1). Cluster analysis based on 46 selected microRNAs (p value ≤0.05; fold change ≥1.5 or ≤0.66) generated a tree with a clear distinction between the preeclampsia and control groups (Fig. 2A).
Hierarchical cluster analysis and validation of differentially expressed microRNAs.
46 differentially expressed microRNAs were selected using the following criteria: q value ≤0.05; fold change ≥1.5 or ≤0.66. The relative expression level was log transformed. Each column represents a placental sample from a pregnant control without preeclampsia (C1–C9) or with preeclampsia (P1–P10). The colour bar in the upper left indicates high expression (red) or low expression (green) (A). Validation of differentially expressed microRNAs by qRT-PCR. 2 down-regulated microRNAs (B) and 8 up-regulated microRNAs (C) are validated by qRT-PCR with independent control placentas (n = 20) and PE placentas (n = 22). The relative expression of each unique miRNA was normalized by the value of the U6 gene. We normalized the control as 1 by dividing each value by the average value of the control group. PE/Control–HTS indicates the fold change of PE placental microRNA expression level relative to the control as demonstrated by HTS. The mean raw ΔCT (CT,miRNA − CT,U6) values of each miRNA were shown on top of the bars. Data are shown as the mean ± S.E.M. Significant differences were determined by 2-tailed Student’s t test or the Mann-Whitney test. ***P < 0.001, **P < 0.01, *P < 0.05.
Validation of differentially expressed microRNAs by qRT-PCR
Quantitative RT-PCR was carried out to validate the differentially expressed miRNAs in preeclampsia detected by HTS (Fig. 2B,C). Twelve miRNAs that demonstrated differential expressions in PE by HTS were selected for qRT-PCR validation based on independent 22 PE placentas and 20 control placentas. Ten miRNAs were successfully validated by qRT-PCR, including 8 up-regulated miRNAs (miR-148a-3p, miR-210, miR-193b-3p, miR-31-5p, miR-365a-3p, miR-516b-5p, miR-520a-5p and miR-27a-5p) and 2 down-regulated miRNAs (miR-135b-5p and miR-136-3p). Additional 2 miRNAs (miR-10b-5p and miR-192-5p) were not validated in these independent samples.
Consistency of the identified differentially expressed microRNAs between the present study and previous reports
Using a combination of the differentially expressed microRNAs from our study and the results based on previous array analysis, we identified a complete list of 182 differentially expressed miRNAs. Forty-four differentially expressed miRNAs are present in Supplementary Table S2. These miRNAs were identified in at least two studies and most of the studies on the corresponding miRNAs reported consistent findings. MiR-210, which was reported in 8 studies, was the most common recurring miRNA among the 44 differentially expressed miRNAs. Indeed, the function of miR-210 in preeclampsia has also been widely investigated compared to other miRNAs12,22,23. The second most common recurring miRNA was miR-193b-3p, which was presented in 4 studies and consistently showed up-regulation in preeclampsia. However, to our knowledge, no research on the function of miR-193b-3p in preeclampsia has been conducted to date. Although there are additional 2 miRNAs (miR-181a-5p and miR-363-3p) present in 4 studies, they were not found to be differentially expressed in our study. Focused on our own study, 8 differentially expressed miRNAs (miR-210, miR-193b-3p, miR-31-5p, miR-193b-5p, miR-10b-5p, miR-151-3p, miR-520-3p and miR-584-5p) were found to be consistent with at least one previous study. One miRNA, miR-192-5p, which demonstrated differential expression in our HTS analysis, was also present in two previous studies with the opposite findings. It should be noted that miR-192-5p showed no differential expression in our qRT-PCR validation in an independent cohort. The remaining differentially expressed miRNAs (16 miRNAs) from our study were newly found.
Effect of miR-193b-3p on HTR-8/SVneo migration and invasion
We focused on microRNAs that demonstrated consistent findings among a number of independent studies. MiR-193b-3p was the second most common recurring miRNA after miR-210. We inferred that miR-193b-3p might have an important role in the progression of preeclampsia.
We found that the genes predicted to be targets of miR-193b-3p by TargetScan were enriched in the following gene ontology categories: cell motility and migration, cell proliferation, positive regulation of nitrogen compound metabolic process and angiogenesis (Fig. 3A). In addition, we noticed that the most significantly enriched functional annotation cluster among these 221 targets of miR-193b-3p was cell motility, cell migration and cell motion (Fig. 3B). This result suggests that miR-193b-3p directly or indirectly regulates genes involved in cell migration.
Gene ontology (GO) analysis of predicted targets of miR-193b-3p.
Gene ontology analysis of predicted 221 targets of miR-193b-3p (predicted by TargetScan) was performed on DAVID (A). The most significantly enriched functional annotation cluster of the GO analysis is cell motility, cell migration and cell motion (B).
We then evaluated the effects of miR-193b-3p on the migration of a trophoblast (HTR-8/SVneo) cell line with in vitro scratch assay (Fig. 4B) and transwell migration assay (Fig. 4C). We evaluated the efficiency of miR-193b-3p overexpression constructs (Pre-miR-193b-3p) and miR-193b-3p inhibitor (Fig. 4A). The fold-change of relative miR-193b-3p expression after transfecting Pre-miR-193b-3p in HTR-8/SVneo was 2.54. This fold-change and the relative miR-193b-3p expression in HTR-8/SVneo were similar to the result from our miRNA profiling of PE and control placentas by HTS. Transfecting miR-193b-3p inhibitor decreased the expression of miR-193b-3p in HTR-8/SVneo by 95%. As shown in our in vitro scratch assay, miR-193b-3p significantly decreased the migration of HTR-8/SVneo by 36% compared to the corresponding negative control (P < 0.001, Fig. 4B) and inhibition of miR-193b-3p significantly improved the migration of HTR-8/SVneo by 15% compared to the corresponding negative control (P < 0.05, Fig. 4B). As shown in transwell migration assay, miR-193b-3p significantly decreased the migration of HTR-8/SVneo by 37% compared to the corresponding negative control (P < 0.001, Fig. 4C) and inhibition of miR-193b-3p significantly improved the migration of HTR-8/SVneo by 19% compared to the corresponding negative control (P < 0.01, Fig. 4C).
Function study of miR-193b-3p in preeclampsia.
We evaluated the efficiency of Pre-miR-193b-3p and miR-193b-3p inhibitor (A). Trophoblast (HTR-8/SVneo) cells were transiently transfected with pre-miR-193b-3p, miR-193b-3p inhibitor or the corresponding control constructs (negative control, NC). The migration ability was assessed 24 h from the scratch (B). To investigate the effect of miR-193b-3p overexpression, 54 scratch spots were assessed for either Pre-miR-193b-3p group or NC pre-miR-193b-3p group. To investigate the effect of miR-193b-3p inhibition, 10 scratch spots were assessed for each experiment group. The transwell migration assay was assessed 6 h from plating cells (C). The transwell invasion assay was assessed 24 h from plating cells (D). The number of invaded cells was counted in 5 different fields of each membrane under light microscope. All experiments were repeated three times. Data are shown as the mean ± SEM. Significant differences were determined by Student’s t test. ***P < 0.001, **P < 0.01.
We also assessed the effects of miR-193b-3p on the invasion of a trophoblast (HTR-8/SVneo) cell line with transwell invasion assay. MiR-193b-3p significantly decreased the invasion of HTR-8/SVneo by 41% compared to the corresponding negative control (P < 0.001, Fig. 4D) and inhibition of miR-193b-3p significantly improved the invasion of HTR-8/SVneo by 88% compared to the corresponding negative control (P < 0.001, Fig. 4D).
TGF-β2 was identified as a direct target of miR-193b-3p
We re-investigated the results of our previous study concerning protein-coding gene expression profiles in PE placentas24. We found that TGF-β2 was the only differentially expressed gene among the 16 predicted miR-193b-3p targets involved in the functional annotation cluster of cell motility, cell migration and cell motion (Fig. 3B).
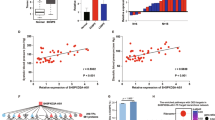
To analyse whether miR-193b-3p directly binds to the predicted conserved binding site, we cloned the full length of TGF-β2 3′UTR into the pGL3 Luciferase Reporter Vector (pGL3-TGF-β2) and the same TGF-β2 3′UTR with a mutation at the corresponding conserved miR-193b-3p binding site was also cloned into the pGL3 Luciferase Reporter Vector (pGL3-TGF-β2 mutation). Relative luciferase activity of constructs with wild type TGF-β2 3′UTR in 293 cell line was reduced to 65% (P < 0.001, Fig. 5A) when overexpressing miR-193b-3p, and was increased to 164% (P < 0.05, Fig. 5A) when inhibiting miR-193b-3p; whereas a change in luciferase activity was not observed in the construct with mutant TGF-β2 3′UTR. Meanwhile, relative luciferase activity of constructs with wild type TGF-β2 3′UTR in HTR-8/SVneo was reduced to 60% (P < 0.001, Fig. 5A) when overexpressing miR-193b-3p and was increased to 141% (P < 0.01, Fig. 5A) when inhibiting miR-193b-3p; whereas a change in luciferase activity was not observed in the construct with mutant TGF-β2 3′UTR. We confirmed that miR-193b-3p could repress TGF-β2 expression by directly binding to a conserved 3′UTR site of TGF-β2.
Validation of TGF-β2 as the direct target of miR-193b-3p.
The pGL3 Luciferase Reporter Vector (pGL3-TGF-β2 or pGL3-TGF-β2 mutation) were cotransfected with the miR-193b-3p precursor (Pre-miR-193b-3p), miR-193b-3p Inhibitor or the corresponding control constructs (negative control, NC) into 293 and trophoblast (HTR-8/SVneo) cell lines. Luciferase activity was measured (A). Sequence alignment between the normal and mutated sequence of TGF-β2 3′-UTR and miR-193b-3p was showed in the lower channel. When constructing the mutation type, 7 bp of the wild TGFβ2 3′UTR which matched the seed sequence of miR-193b-3p was deleted. (A). HTR-8/SVneo cells were transfected with the miR-193b-3p precursor (Pre-miR-193b-3p), miR-193b-3p Inhibitor or the corresponding control constructs (negative control, NC) and the TGF-β2 mRNA level was assessed (B). TGF-β2 mRNA was significantly down-regulated in PE placentas (C) and was negatively correlated with the expression of miR-193b-3p (D).
We also assessed the TGF-β2 mRNA level in HTR-8/SVneo cells after transfection with miR-193b-3p precursor (Pre-miR-193b-3p), miR-193b-3p Inhibitor or control constructs (negative control, NC). miR-193b-3p significantly decreased TGF-β2 mRNA level in HTR-8/SVneo cells by 64% compared to the corresponding negative control (P < 0.001, Fig. 5B) and inhibition of miR-193b-3p significantly improved the expression of TGF-β2 mRNA level in HTR-8/SVneo by 23% compared to the corresponding negative control (P < 0.001, Fig. 5B).
We then measured TGF-β2 levels by qRT-PCR in all placental tissues and found that TGF-β2 mRNA was significantly down-regulated in PE placentas (P < 0.01, Fig. 5C). A significant negative correlation between the expression of miR-193b-3p and TGF-β2 mRNA was observed (P < 0.001, Fig. 5D). We also found that the TGF-β2 protein level was approximately 65% lower in PE placentas (Supplementary Fig. S1).
The function of miR-193b-3p was exerted via regulation of TGF-β2
To verify whether the function of miR-193b-3p would be exerted via regulation of TGF-β2, we determined whether it could be possible to rescue miR-193b-3p inhibiting effect by TGF-β2 overexpression in HTR8/SVneo cells. Overexpression of TGF-β2 partially attenuated the inhibitory effects of miR-193b-3p on cell migration (P = 0.025, Fig. 6B) and cell invasion (P < 0.01, Fig. 6B) at 48 h after transfection.
Restoring TGF-β2 expression reverses the regulation of miR-193b-3p on Trophoblast (HTR-8/SVneo) cells migration and invasion.
Simultaneous transfection of HTR8/SVneo cells with the miR-193b-3p inhibitor and TGF-β2 siRNA reduced the ability of the miR-193b-3p inhibitor to upregulate TGF-β2 mRNA expression at 48 h after transfection and co-transfection of pcDNA-TGFβ2 restored TGF-β2 expression (A). Overexpression of TGF-β2 (pcDNA-TGFβ2) attenuates the functions of miR-193b-3p to inhibit cell migration and cell invasion at 48 h after transfection (B). Co-transfection of TGF-β2 siRNA reduced the ability of miR-193b-3p inhibitor to promote cell migration at 48 h after transfection (C). Co-transfection of TGF-β2 siRNA abolished the increased cell invasion promoted by transfection of the miR-193b-3p inhibitor at 48 h (C).
In addition, simultaneous transfection of HTR8/SVneo cells with the miR-193b-3p inhibitor and TGF-β2 siRNA reduced the ability of the miR-193b-3p inhibitor to upregulate TGF-β2 mRNA expression at 48 h after transfection (P < 0.01, Fig. 6A). Furthermore, the transwell migration assay revealed that co-transfection of TGF-β2 siRNA significantly reduced the ability of the miR-193b-3p inhibitor to promote cell migration at 48 h after transfection (P < 0.01, Fig. 6B). Additionally, the transwell invasion assay demonstrated that co-transfection of TGF-β2 siRNA abolished the increased cell invasion promoted by transfection of the miR-193b-3p inhibitor at 48 h (P < 0.001, Fig. 6C). These findings indicated that miR-193b-3p could suppress HTR8/SVneo trophoblast cell migration and invasion through regulating TGF-β2.
Discussion
In the present study, we detected a list of differentially expressed microRNAs in PE placentas by HTS. Eight differentially expressed miRNAs from our study were found to be consistent with at least one previous study and 16 microRNAs were newfound. However, only limited number of miRNAs demonstrated consistent results among different research studies. The inconsistency might be caused by the different clinical characteristics of patients enrolled in these different studies. This also might be due to the application of different experiment platforms and different sample sizes18,25. Although our sample size is relatively larger than previously published studies on the same topic, the small cohort may still be a limitation of the present study. Different experiment platforms have apparent differences in performance in reproducibility, accuracy, detection sensitivity and specificity18. Our follow-up study was focused on miRNAs that demonstrated consistent results among a number of independent studies.
For instance, miR-210 is the most commonly identified differentially expressed miRNA in placental. Indeed, miR-210 is one of the most investigated miRNAs in PE. It is a hypoxia-responsive miRNA involved in trophoblast migration and invasion, siderosis of interstitial trophoblasts and placental mitochondrial dysfunction22,26,27. Up-regulation of placental miR-210 was also detected in our research.
MiR-193b-3p is the second most common recurring miRNA after miR-210. The findings in 3 previous studies and ours are consistent, demonstrating that miR-193b-3p is up-regulated in preeclamptic placentas. Previous publications suggested that miR-193b-3p was a putative tumor suppressor. This gene was found to be greatly down-regulated in several types of malignant tumors28,29. Moreover, overexpression of miR-193b-3p repressed cell proliferation and inhibited cancer cells invasion, migration and growth29,30. Typically, this was reported in the tumour microenvironment, but what about the placental environment? During placental development, cytotrophoblasts proliferate to form anchoring villi and give rise to interstitial trophoblasts that invade the uterine decidua and endovascular trophoblasts that migrate into the maternal spiral arteries; accordingly, the trophoblasts’ capacity for proliferation, migration and invasion plays a crucial role in successful placental development31. It appears that trophoblasts and cancer cells share similar biological characteristics in their proliferative, migratory and invasive properties32,33. Adequate invasion of tumour cells leads to cancer, but inadequate invasion of trophoblasts might result in poor placentation, leading to preeclampsia34. Consequently, the tumour suppressor role of miR-193b-3p is suggestive of its crucial role in the regulation of some principal trophoblasts events, leading to preeclampsia. In silico target prediction of miR-193b-3p by TargetScan and the following GO analysis showed that targets genes of miR-193b-3p were enriched in cell motility and cell migration. In addition, our in vitro scratch assay and transwell migration and invasion assays demonstrated that overexpression of miR-193b-3p significantly decreased the migration and invasion of HTR-8/SVneo cells, while inhibition of miR-193b-3p significantly improved the migration and invasion of HTR-8/SVneo cells. Our results indicated that the aberrant expression of miR-193b-3p in the preeclamptic placenta could contribute to the pathogenesis of preeclampsia through its inhibitory effect on the migration and invasion of trophoblasts.
We also showed that transforming growth factor-beta 2 (TGF-β2), a member of the transforming growth factor-β (TGFβ) superfamily, was post-transcriptionally dysregulated by miR-193b-3p. And restoring TGF-β2 expression reversed the regulation of miR-193b-3p on trophoblast (HTR-8/SVneo) cells migration and invasion. Meanwhile, TGF-β2 mRNA and protein expression were significantly decreased in preeclamptic placentas. According to previous research, the TGFβ signalling pathway is crucial to placental biological processes35. It is involved in regulating a number of cellular processes, including growth, migration, invasion, epithelial to mesenchymal transition (EMT) and immune reactions36,37. For instance, the cells undergoing EMT manifest a migratory phenotype and acquire invasive properties38 and the differentiation of interstitial trophoblasts and endovascular trophoblasts is considered to be an EMT-like transformation process39. Previous studies suggest that TGFβ is potent inducer of EMT in various cells40,41 and showed that the TGFβ family induced the invasive properties of a trophoblast cell line in vitro42. And one study showed that cell migration was improved by inducing TGF-β2 expression under hypoxia43. In addition, soluble endoglin (sEng), which is well known to be involved in preeclampsia, was also identified as a TGFβ inhibitor in previous studies44,45. Therefore, we inferred that aberrant TGFβ signalling in the placenta might play a key role in the pathogenesis of PE; aberrant expression of miR-193b-3p in preeclamptic placentas could exert an inhibitory effect on the migration and invasion of trophoblasts by directly targeting TGF-β2. The detailed mechanism remains to be elucidated.
In conclusion, we detected and verified a list of differentially expressed microRNAs in PE placentas by HTS and qRT-PCR and provided preliminary evidence for the role of miR-193b-3p in the pathogenesis of preeclampsia by targeting TGF-β2.
Methods
Sample collection
All placenta samples used in this study were collected from normal pregnant woman who were Han Chinese in origin (control group; n = 29) and from preeclamptic pregnant patients (PE group; n = 31) by elective caesarean delivery in the absence of labour during the third trimester of gestation (Table 2, All results are presented as the mean ± S.D.; BMI, indicates body mass index; DBP, diastolic blood pressure; and SBP, systolic blood pressure). Detailed clinical characteristics of patients with 9 normal and 9 preeclamptic pregnancies in HTS study were shown in Supplementary Table S3. PE was defined as systolic blood pressure (SBP) ≥140 mmHg and/or diastolic blood pressure (DBP) ≥90 mmHg on 2 occasions at least 4 hours apart with proteinuria ≥300 mg/day from 24 h urine collection occurring after the 20th week of gestation but resolving by the 12th week postpartum. All clinical placentas were collected immediately after the caesarean section. The placental samples at the chorionic plate were separately taken from each quadrants along with central portion in the placenta disc. ~1 cm3 fragments were dissected from the placenta, after removal of maternal blood by vigorous washing in phosphate buffered saline (PBS). The tissues were maintained in centrifuge tubes with RNAlater (Ambion Inc., Austin, TX) and then frozen at −80 °C. All the patients did not accepte antihypertensive medication and other special medical treatment before termination of pregnancy. All women enrolled in the study gave their written informed consent to the sample collection and analyses. This research was approved by the Institutional Review Committee of Institutes of Biomedical Sciences at Fudan University. All experiments were performed in accordance with relevant guidelines and regulations.
MiRNA isolation and sequencing analysis
Total RNA was extracted using the Ambion mirVana™PARIS™Kit (Ambion , Carlsbad, California, USA) according to the manufacturer’s instruction. RNA concentration and quality were determined with Agilent 2100 Bioanalyser (Agilent, Palo Alto, CA). 1 ug total RNA was obtained for the small RNA sequencing analysis and underwent further processing with the TruSeq Small RNA Sample Preparation Kit. Briefly, 1 ug total RNA underwent adapter ligation, reverse transcription, PCR amplification and pooled gel purification to generate a library product. Following this, we performed microRNA-sequencing using the Illumina HiSeq 2000 sequencing system with approximately 6 multiplexed samples in each sequencing lane. The raw Illumina reads were pre-processed and mapped. The number of mapped reads was an average of 25.8 million in the control group and 28.4 million in the PE group. Using R version 2.14.0 statistical software, we screened the differentially expressed microRNAs based on the following criteria: p value ≤0.01; fold change ≥1.5 or ≤0.66.
Validation of mRNA and miRNA by qRT-PCR
A total of 500 ng RNA was isolated for qRT-PCR validation. We performed real-time quantification of miRNAs by stem–loop RT–PCR based on previous studies46. The primers used are shown in supplementary table S4. Briefly, reverse transcriptase reactions were conducted with a total of 12.5 ul comprised of RNA samples, stem–loop RT primer, AMV Reverse Transcriptase (Promega, Madison, Wisconsin, USA), 5 × RT buffer (Promega, Madison, Wisconsin, USA), dNTPs (Promega, Madison, Wisconsin, USA) and RNase inhibitor (Promega, Madison, Wisconsin, USA). Real-time PCR was performed with samples containing RT product, microRNA-specific forward primer, universal reverse primer, ProbeLibrary Probe #21 (Roche, Penzberg, Germany) and FastStart Universal Probe Master (Rox) (Roche, Penzberg, Germany) using the Applied Biosystems 7900HT PCR system. RNU6B (U6) was used to normalize the microRNA qRT-PCR data.
A total of 500 ng RNA was isolated for mRNA qRT-PCR. The cDNA was synthesized using AMV Reverse Transcriptase system (Promega, Madison, Wisconsin, USA). TGF-β2 mRNA qRT-PCR was performed using the Applied Biosystems 7900HT with FastStart Universal SYBR Green Master (Rox) (Roche, Penzberg, Germany). The primers for TGF-β2 mRNA qRT-PCR are described in Supplementary Table S4.
The qRT-PCR data was analysed using the ΔΔCt method.
Prediction of miR-193b-3p targets and GO analysis
We predicted targets of miR-193b-3p with only evolutionary conserved sites by TargetScan (TargetScanHuman 6.2) and 221 targets of miR-193b-3p were included. And then, we performed gene ontology(GO) analysis by use of DAVID (http://david.abcc.ncifcrf.gov/).
Literature Review
We performed a search of literature published to October 2014 through PubMed to identify all articles reporting the differential expression of miRNAs in preeclampsia. In the 80 identified studies by using the the keywords “microrna” and “preeclampsia”, 33 were studies of human preeclamptic placentas compared to normotensive controls, which included 12 array studies based on hybridization, RT-qPCR or sequencing analysis that presented the results as a list of differentially expressed miRNAs10,11,12,13,14,15,19,21,47,48,49,50,51,52,53 and 21 studies that focused on specific miRNAs.
Plasmid construction
A 119 base-pair DNA fragment encompassing the has-mir-193b gene was amplified with PCR from genomic DNA by use of the following primers: forward, 5′-TCTCCAAACTCTTGCCTCAAAG-3′ and reverse, 5′-CCAGCCGGGTTTTGGACG-3′. The products were cloned into the pSilencer 4.1 cmv puro vector (Applied Biosystems, Waltham, Massachusetts, USA) between the BamH I and Hind III sites as pre-miR-193b-3p.
The whole length of human TGF-β2 3′UTR was amplified with the following primers: forward, 5′-CTAGCTAGCAATTCTTGGAAAAGTGGCAAGACC-3′and reverse, 5′-CTAGCTAGCACTAGACAAAGCAAGACAACCAGA-3′. The products were cloned into the pGL3 promoter vector (Promega, Madison, Wisconsin, USA) downstream of the Luciferase reporter gene in the Nhe I site as pGL3-TGF-β2.
Expression plasmids of pGL3-TGF-β2-mutation without the miR-193b-3p binding site(s) were constructed by sequence overlapped extension-PCR with the following mutation primer sequences: forward, 5′-CTTTTGAGTAAAGCCCCTATA-3′ and reverse, 5′-CATACATTTGTGAGTGATCATTA-3′. A 1,406-bp fragment of the TGF-β2 CDS region was amplified with PCR from the cDNA of HTR-8/SVneo cell line using primers in Supplementary Table S4. The PCR product was cloned into a pcDNA (Applied Biosystems, Waltham, Massachusetts, USA) using Xba I and Xho I restriction sites as the TGF-β2 expression vector pcDNA-TGFβ2.
Cell culture and transfection
To investigate the effect of miR-193b-3p overexpression, HTR-8/SVneo cells were cultured in RPMI-1640 (Gibco, California, USA) on a 24-well plate and transfected with 800 ng of pre-miR-193b-3p plasmid or control constructs (negative control, NC pre-miR-193b-3p) per well. To investigate the effect of miR-193b-3p Inhibition, HTR-8/SVneo cells were cultured in RPMI-1640 (Gibco, California, USA) on a 24-well plate and transfected with 100 pmol miR-193b-3p inhibitor (GenePharma, China) or control constructs (negative control, NC Inhibitor) per well. To investigate the roles of TGF-β2 during the suppression of migration and invasion of trophoblast cells by miR-193b-3p, we co-transfected HTR8/SVneo cells with 200 ng pre-miR-193b-3p and 200 ng pcDNA-TGFβ2 to perform a rescue experiment. We also co-transfected HTR8/SVneo cells with 50 pmol miR-193b-3p inhibitor (GenePharma, China) and 50 pmol TGF-β2 siRNA (GenePharma, China) to perform a rescue experiment. The sequence information of miR-193b-3p inhibitor (GenePharma, China) and TGF-β2 siRNA (GenePharma, China) were described in Supplementary Table S4.
In vitro scratch assay
An in vitro scratch assay was used as described previously54. The protocol of cell culture and transient transfection is described previously. When HTR-8/SVneo cells were grown to 90% confluence, the scratches were made. In order to minimize cell proliferation, the cells were allowed to grow in RPMI-1640 (Gibco, California, USA) without foetal bovine serum. The width of the scratch was monitored by LEICA DMI4000B within 24 h after transfection.
Transwell migration assay
The protocol of cell culture and transient transfection is described previously. Transwell compartments were prepared with 24-well format and 8 μm pore size insert. For the lower compartment, we added 0.8 ml of RPMI-1640 (Gibco, California, USA) with 10% FBS; and for the upper compartment, we gently added 5 × 104 cells in 100 μl serum-free RPMI-1640 (Gibco, California, USA). We incubated the cells in the transwell plate at 37 °C and 5% CO2 for 6 h. After 6 h, we carefully took the insert out. Cells on the upper surface of membranes were completely removed. and the cells migrated to the lower surface of membranes were fixed with 100% formaldehyde and stained with 0.05% crystal violet. The number of migrated cells was counted under LEICA DMI4000B. Cell migration level was presented as the percentage of migrated cell number compared with the corresponding control.
Transwell invasion assay
The protocol of cell culture and transient transfection was described previously. We prepared for the transwell 24-well fitted inserts (8 μm pore size, Millipore Corp, Massachusetts, USA). The transwell inserts were pre-coated with 7 μg matrigel (BD Biosciences, NJ, USA) in 50 μl serum-free culture medium. For the lower compartment, we added 0.8 ml of RPMI-1640 (Gibco, California, USA) with 10% FBS. For the upper compartment, we gently added 5 × 104 cells in 100 μl serum-free RPMI-1640 (Gibco, California, USA). We incubated the cells in the transwell plate at 37 °C and 5% CO2 for 24 h. After 24 h, we carefully took the insert out. Cells on the upper surface of membranes were completely removed and the cells migrated to the lower surface of membranes were fixed with 100% formaldehyde and stained with 0.05% crystal violet. The number of invaded cells was counted under LEICA DMI4000B. Cell invasion level was presented as the percentage of invaded cell number compared with the corresponding control.
Luciferase assays
Luciferase assays were performed with the Dual-Luciferase Reporter Assay System (Promega, Madison, Wisconsin, USA). pRL-SV40 (Promega, Madison, Wisconsin, USA), a control plasmid, was used to check for transfection efficiency. To investigate the effect of miR-193b-3p overexpression, 293 and HTR-8/SVneo cells were cotransfected with luciferase reporter plasmids (300 ng), pRL-SV40 (15 ng) and pre-miR-193b-3p expression vector (500 ng) or control constructs NC pre-miR-193b-3p (500 ng) per well. To investigate the effect of miR-193b-3p inhibition, 293 and HTR-8/SVneo cells were cotransfected with luciferase reporter plasmids (300 ng), pRL-SV40 (15 ng) and miR-193b-3p Inhibitor (100 pmol) or control NC Inhibitor (100 pmol).
Western blotting
Placental tissues were homogenized in lysis buffer with added proteinase inhibitors (Roche, Penzberg, Germany). A BCA kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA) was used to assess protein concentrations. 20 ug of proteins were resolved by 10% SDS-PAGE and transferred to a nitrocellulose membrane (Applied Biosystems, Waltham, Massachusetts, USA). The membranes were incubated with the primary antibodies against TGF-β2 (Abcam, Cambridge, UK, 1:500) and GAPDH (Abmart, Shanghai, China, 1:1000) overnight at 4 °C. Following this, the membranes were incubated with horseradish peroxidase (HRP) conjugated secondary antibodies (Abmart, Shanghai, China, 1:3000). The signal intensity was quantitatively assessed with Fujifilm LAS-3000 Imager and was normalized to GAPDH.
Statistical analysis
Analysis of microRNA-seq data was performed using R version 2.13.1. A 2-tailed t test was performed; and Mann-Whitney test was performed with SPSS 17 (SPSS Inc., Chicago, Illinois, USA) when the data pattern did not follow Gaussian distribution. The results are shown as the mean ± S.E.M. or mean ± S.D. A threshold of P value <0.05 was considered to be significant. Correlation was calculated using Pearson correlation coefficient.
Additional Information
How to cite this article: Zhou, X. et al. The aberrantly expressed miR-193b-3p contributes to preeclampsia through regulating transforming growth factor-β signalling. Sci. Rep. 6, 19910; doi: 10.1038/srep19910 (2016).
References
Steegers, E. A., von Dadelszen, P., Duvekot, J. J. & Pijnenborg, R. Pre-eclampsia. Lancet. 376, 631–644 (2010).
Duley, L. The global impact of pre-eclampsia and eclampsia. Semin Perinatol. 33, 130–137 (2009).
Acosta-Sison, H. The relationship of hydatidiform mole to pre-eclampsia and eclampsia; a study of 85 cases. Am J Obstet Gynecol. 71, 1279–1282 (1956).
Redman, C. W. & Sargent, I. L. Immunology of pre-eclampsia. Am J Reprod Immunol. 63, 534–543 (2010).
Huang, S. J. et al. The implication of aberrant GM-CSF expression in decidual cells in the pathogenesis of preeclampsia. Am J Pathol. 177, 2472–2482 (2010).
Paiva, P., Salamonsen, L. A., Manuelpillai, U. & Dimitriadis, E. Interleukin 11 inhibits human trophoblast invasion indicating a likely role in the decidual restraint of trophoblast invasion during placentation. Biol Reprod. 80, 302–310 (2009).
Chaiworapongsa, T., Chaemsaithong, P., Yeo, L. & Romero, R. Pre-eclampsia part 1: current understanding of its pathophysiology. Nat Rev Nephrol. 10, 466–480 (2014).
Ambros, V. The functions of animal microRNAs. Nature. 431, 350–355 (2004).
Fu, G., Brkic, J., Hayder, H. & Peng, C. MicroRNAs in human placental development and pregnancy complications. Int J Mol Sci. 14, 5519–5544 (2013).
Xu, P. et al. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension. 63, 1276–1284 (2014).
Betoni, J. S. et al. MicroRNA analysis in placentas from patients with preeclampsia: comparison of new and published results. Hypertens Pregnancy. 32, 321–339 (2013).
Ishibashi, O. et al. Hydroxysteroid (17-beta) dehydrogenase 1 is dysregulated by miR-210 and miR-518c that are aberrantly expressed in preeclamptic placentas: a novel marker for predicting preeclampsia. Hypertension. 59, 265–273 (2012).
Enquobahrie, D. A. et al. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 204, 178 e112–121 (2011).
Zhu, X. M., Han, T., Sargent, I. L., Yin, G. W. & Yao, Y. Q. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 200, 661 e661–667 (2009).
Pineles, B. L. et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 196, 261 (2007).
Dai, Y. et al. MicroRNA-155 inhibits proliferation and migration of human extravillous trophoblast derived HTR-8/SVneo cells via down-regulating cyclin D1. Placenta. 33, 824–829 (2012).
Luo, L. et al. MicroRNA-378a-5p promotes trophoblast cell survival, migration and invasion by targeting Nodal. J Cell Sci. 125, 3124–3132 (2012).
Mestdagh, P. et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 11, 809–815 (2014).
Guo, L. et al. A comprehensive survey of miRNA repertoire and 3′ addition events in the placentas of patients with pre-eclampsia from high-throughput sequencing. PloS one. 6 (2011).
Weedon-Fekjaer, M. S. et al. Placental miR-1301 is dysregulated in early-onset preeclampsia and inversely correlated with maternal circulating leptin. Placenta. 35, 709–717 (2014).
Yang, S., Li, H., Ge, Q., Guo, L. & Chen, F. Deregulated microRNA species in the plasma and placenta of patients with preeclampsia. Mol Med Rep. 12, 527–534 (2015).
Lee, D. C. et al. miR-210 targets iron-sulfur cluster scaffold homologue in human trophoblast cell lines: siderosis of interstitial trophoblasts as a novel pathology of preterm preeclampsia and small-for-gestational-age pregnancies. Am J Pathol. 179, 590–602 (2011).
Luo, R. et al. MicroRNA-210 contributes to preeclampsia by downregulating potassium channel modulatory factor 1. Hypertension. 64, 839–845 (2014).
Xiang, Y. et al. Up-regulated expression and aberrant DNA methylation of LEP and SH3PXD2A in pre-eclampsia. PloS one. 8 (2013).
Kelly, A. D. et al. Next-generation sequencing and microarray-based interrogation of microRNAs from formalin-fixed, paraffin-embedded tissue: preliminary assessment of cross-platform concordance. Genomics. 102, 8–14 (2013).
Anton, L. et al. miR-210 inhibits trophoblast invasion and is a serum biomarker for preeclampsia. Am J Pathol. 183, 1437–1445 (2013).
Muralimanoharan, S. et al. MIR-210 modulates mitochondrial respiration in placenta with preeclampsia. Placenta. 33, 816–823 (2012).
Chen, J. et al. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 176, 2520–2529 (2010).
Li, X. F., Yan, P. J. & Shao, Z. M. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 28, 3937–3948 (2009).
Gastaldi, C. et al. miR-193b/365a cluster controls progression of epidermal squamous cell carcinoma. Carcinogenesis. 35, 1110–1120 (2014).
Knofler, M. Critical growth factors and signalling pathways controlling human trophoblast invasion. Int J Dev Biol. 54, 269–280 (2010).
Lala, P. K., Lee, B. P., Xu, G. & Chakraborty, C. Human placental trophoblast as an in vitro model for tumor progression. Can J Physiol Pharmacol. 80, 142–149 (2002).
Moslehi, R. et al. Integrative transcriptome analysis reveals dysregulation of canonical cancer molecular pathways in placenta leading to preeclampsia. Sci Rep. 3, 2407 (2013).
Zhou, Y., Damsky, C. H. & Fisher, S. J. Preeclampsia is associated with failure of human cytotrophoblasts to mimic a vascular adhesion phenotype. One cause of defective endovascular invasion in this syndrome? J Clin Invest. 99, 2152–2164 (1997).
Forbes, K., Souquet, B., Garside, R., Aplin, J. D. & Westwood, M. Transforming growth factor-{beta} (TGF{beta}) receptors I/II differentially regulate TGF{beta}1 and IGF-binding protein-3 mitogenic effects in the human placenta. Endocrinology. 151, 1723–1731 (2010).
Lafontaine, L. et al. Transforming growth factor Beta regulates proliferation and invasion of rat placental cell lines. Biol Reprod. 84, 553–559 (2011).
Massague, J. TGFbeta in Cancer. Cell. 134, 215–230 (2008).
Kang, Y. & Massague, J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 118, 277–279 (2004).
Vicovac, L. & Aplin, J. D. Epithelial-mesenchymal transition during trophoblast differentiation. Acta Anat (Basel). 156, 202–216 (1996).
Thiery, J. P. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 15, 740–746 (2003).
Quail, D. F., Zhang, G., Findlay, S. D., Hess, D. A. & Postovit, L. M. Nodal promotes invasive phenotypes via a mitogen-activated protein kinase-dependent pathway. Oncogene. 33, 461–473 (2014).
Law, J., Zhang, G., Dragan, M., Postovit, L. M. & Bhattacharya, M. Nodal signals via beta-arrestins and RalGTPases to regulate trophoblast invasion. Cell Signal. 26, 1935–1942 (2014).
Nagpal, N. et al. HIF-inducible miR-191 promotes migration in breast cancer through complex regulation of TGFbeta-signaling in hypoxic microenvironment. Sci Rep. 5, 9650 (2015).
Li, H. et al. Artificial oxygen carriers rescue placental hypoxia and improve fetal development in the rat pre-eclampsia model. Sci Rep. 16, 15271 (2015).
Venkatesha, S. et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat Med. 12, 642–649 (2006).
Chen, C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179 (2005).
Choi, S. Y. et al. MicroRNA expression profiles in placenta with severe preeclampsia using a PNA-based microarray. Placenta 34, 799–804 (2013).
Hu, Y. et al. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin Chem Lab Med. 47, 923–929 (2009).
Mayor-Lynn, K., Toloubeydokhti, T., Cruz, A. C. & Chegini, N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci. 18, 46–56 (2011).
Noack, F. et al. miRNA expression profiling in formalin-fixed and paraffin-embedded placental tissue samples from pregnancies with severe preeclampsia. J Perinat Med. 39, 267–271 (2011).
Wang, W. et al. Preeclampsia up-regulates angiogenesis-associated microRNA (i.e., miR-17, -20a and -20b) that target ephrin-B2 and EPHB4 in human placenta. J Clin Endocrinol Metab. 97, E1051–1059 (2012).
Zhang, C. et al. Placental miR-106a approximately 363 cluster is dysregulated in preeclamptic placenta. Placenta. 36, 250–252 (2015).
Jiang, F. et al. Upregulation of microRNA-335 and microRNA-584 contributes to the pathogenesis of severe preeclampsia through downregulation of endothelial nitric oxide synthase. Mol Med Rep. 12, 5383–5390 (2015).
Liang, C. C., Park, A. Y. & Guan, J. L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2, 329–333 (2007).
Acknowledgements
We thank Professor Peeyush Lala at the University of Western Ontario, Canada, for the kind gift of HTR-8/SVneo cell line. This work was supported by the National Basic Research Program of China (2015CB943300, 2013CB530740), the Shanghai Municipal Commission of Science and Technology Program (14DJ1400102), the Shanghai Natural Science Foundation (14ZR1401400), the National Natural Science Foundation of China (81501276), the Key Basic Research Foundation of Science and Technology Commission of Shanghai Municipality (13JC1403700), the National Key Technology R&D Program (2012BAI01B09), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning(to Mingqing Xu).
Author information
Authors and Affiliations
Contributions
X.Z. and Q.L. performed the experiments and drafted the manuscript; M.X., L.W. and X.Z. conceived and managed the project; J.X. performed small RNA sequencing analysis; X.Z., H.Z. and Y.X. contributed to experiment design; C.F., T.W., S.X., M.X. and Q.Z. contributed to data analysis; Q.X., M.X. and L.H. revised and edited the manuscript; M.X. and X.Z shared the senior authorship. All authors reviewed the manuscript before submission.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhou, X., Li, Q., Xu, J. et al. The aberrantly expressed miR-193b-3p contributes to preeclampsia through regulating transforming growth factor-β signaling. Sci Rep 6, 19910 (2016). https://doi.org/10.1038/srep19910
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19910
This article is cited by
-
Integrated analysis of microRNA and mRNA expression profiles in Preeclampsia
BMC Medical Genomics (2023)
-
Comparative RNA-seq analysis and ceRNA network of genistein-treated GT1-7 neurons
Molecular & Cellular Toxicology (2023)
-
miR-31-5p from placental and peripheral blood exosomes is a potential biomarker to diagnose preeclampsia
Hereditas (2022)
-
A novel prognostic model for hepatocellular carcinoma based on 5 microRNAs related to vascular invasion
BMC Medical Genomics (2022)
-
Elevated histone demethylase KDM5C increases recurrent miscarriage risk by preventing trophoblast proliferation and invasion
Cell Death Discovery (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.