Abstract
Biofilm formation is a major pathogenicity strategy of Staphylococcus epidermidis causing various medical-device infections. Persister cells have been implicated in treatment failure of such infections. We sought to profile bacterial subpopulations residing in S. epidermidis biofilms and to establish persister-targeting treatment strategies to eradicate biofilms. Population analysis was performed by challenging single biofilm cells with antibiotics at increasing concentrations ranging from planktonic minimum bactericidal concentrations (MBCs) to biofilm MBCs (MBCbiofilm). Two populations of “persister cells” were observed: bacteria that survived antibiotics at MBCbiofilm for 24/48 hours were referred to as dormant cells; those selected with antibiotics at 8 X MICs for 3 hours (excluding dormant cells) were defined as tolerant-but-killable (TBK) cells. Antibiotic regimens targeting dormant cells were tested in vitro for their efficacies in eradicating persister cells and intact biofilms. This study confirmed that there are at least three subpopulations within a S. epidermidis biofilm: normal cells, dormant cells and TBK cells. Biofilms comprise more TBK cells and dormant cells than their log-planktonic counterparts. Using antibiotic regimens targeting dormant cells, i.e. effective antibiotics at MBCbiofilm for an extended period, might eradicate S. epidermidis biofilms. Potential uses for this strategy are in antibiotic lock techniques and inhaled aerosolized antibiotics.
Similar content being viewed by others
Introduction
Bacteria employ a variety of strategies to escape killing by antibiotics, including mutation, phenotypic variation and change to a biofilm growth mode1,2,3. One form of phenotypic variation, known as persistence, is characterised by the presence of a subset of antibiotic-tolerant cells within a bacterial population. Persister cells pre-exist in most bacterial populations, including cultures at mid-log phase, stationary phase and in biofilms4,5,6. The prevalence of persister cells in a population depends on its growth mode, the age of inocula, strain background, growth medium and time course chosen for selection6,7,8. Persister cells display heterogeneity in growth rates and tolerance to various antibiotics6,9,10, though multidrug tolerance is not a consistent trait11. Transcriptome analysis suggests that persister cells have reduced expression of genes involved in metabolic pathways, biosynthesis pathways and energy production12,13, which often leads to a dormancy status of cells. However, being entirely dormant is not necessarily a prerequisite for the formation of persister subpopulations14,15,16.
In clinical settings, bacteria grow predominantly as biofilms, following attachment and accumulation on biotic or abiotic surfaces; and they present group dynamics17,18,19. Bacteria within biofilms are highly tolerant to antibiotics, but the exact mechanisms behind this tolerance are complex and no single factor can fully account for this specific trait20,21,22. Persister cells are more prevalent in biofilms than in log-planktonic cultures and are thought to be responsible for the recalcitrance of many chronic infections, such as cystic fibrosis and chronic wound infections, to antibiotic treatment1,3,9,23. Although the role of persister cells in biofilm drug-tolerance has been reported by several studies24,25,26, quantitative evidence to support this is limited by the technical difficulty in excluding other biofilm-related factors when isolating persister cells. These include the presence of extracellular polymer substances (EPS), quorum-sensing (QS) factors and extracellular hydrolytic enzymes that may also protect the non-persister cells from the action of antibiotics27,28,29,30,31. Furthermore, the reported proportions of persister cells in bacterial biofilms varies, even when biofilms were cultured under similar in vitro conditions32. For instance, Shapiro et al. reported that the percentages of persister cells isolated from a S. epidermidis RP62A biofilm using levofloxacin or vancomycin were 28% and 94% respectively33. In contrast, a well-accepted relapsing biofilm infection model described by Lewis et al. proposed that biofilms comprised only ~0.1–1% persister cells4,9,34. This difference cannot be explained by stochasticity of the persister cell production in a bacterial population, but could be due to different bacterial strains used, or most likely different methodologies used to select persister cells4,20,32,33,35,36. Some researchers isolated biofilm persister cells by challenging the entire biofilm with antibiotics and did not take into consideration the influence of those biofilm-related factors on antibiotic activity33,37,38,39. Others dissociated biofilm cells with sonication and/or vortexing, then challenged them with antibiotics to select persister cells40; this procedure however produced a relatively high proportion of bacterial clumps, which might present attributes resembling those of an intact biofilm. Many previous studies used antibiotics at 8–100 times minimum inhibitory concentration (MIC) and treatment periods between 2.5 hours (h) to 24 h to isolate persister cells6,8,41. Though these methods have been validated to select the generally defined “persister cells”, insufficient killing of all vulnerable cells in a bacterial population has been reported33,42,43. To obtain comparable data on the prevalence of persister cells within bacterial biofilms, an ideal method would require the separation of individual cells from other biofilm-related factors that affect the performance of antibiotics. Such a method should also specify concentrations of antibiotics and exposure times needed to effectively eradicate non-tolerant cells.
We have previously proposed the existence of three cell subpopulations residing within S. epidermidis biofilms; normal cells that are rapidly killed by antibiotics, tolerant-but-killable (TBK) cells that only respond to high concentrations of antibiotics and dormant cells that resist very high concentrations of antibiotics; the latter two comprise persister cells35. In this study, we aimed to provide further support for our previous proposal, by examining the proportions of TBK and dormant cells in S. epidermidis biofilms. The specific aim was to accomplish two tasks in persister cell research recently raised by Lewis et al.9, but specifically related to S. epidermidis biofilms: 1) to detail the role of persister cells in antibiotic tolerance of biofilms; 2) to examine the possibility of eradication of persister cell and biofilms using conventional antibiotics in vitro.
Results
Validation of a method to isolate single cell populations from biofilms
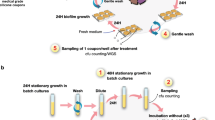
Three-dimensional reconstruction of confocal laser scanning microscopy (CLSM; see supplementary information for experimental details) images showed a large number of survivors residing in biofilms after exposure to vancomycin and oxacillin at MBCbiofilm for 24 h (Fig. 1A); the percentage of survivor cells is inconsistent with that of persister cells in biofilms proposed by other researchers and ourselves3,34,35. The modified broth recovery-based biofilm MBC defined in our previous study was referred to as MBCbiofilm in this study35 (Table 1). This concentration of antibiotics was expected to eradicate normal cells and also “slow-replicating” cells residing in staphylococcal biofilms, leaving only very few viable persister cells35. We reasoned that dissociating biofilms and recovering single-cell population would provide a better measure of the proportion of persister cells than using intact biofilms. After mechanical disruption of S. epidermidis RP62A biofilms by scraping, pipetting, sonication and vortexing and passing the acquired cell suspensions through a 1.2 μm syringe filter, mostly single cells and diplococci were observed by SEM (Fig. 1B). In contrast, bacterial clumps consisting of more than 10 cells were frequently observed when the filtration step was not included (Fig. 1B). Viable counts performed on the bacterial suspensions obtained with or without filtration showed that approximately 45–65% of bacteria in the original suspension passed through the filter (Fig. 1C). GFP-expressing S. epidermidis RP62A cells were constructed and used in flow cytometry (see supplementary information) to measure levels of active and dormant subpopulations of biofilm-embedded cells. Biofilm cell suspensions collected with or without filtration showed similar percentage of GFP + (active) cells (70.9% versus 71.9%), suggesting that these ~45–65% cells are representative of the total population of embedded cells in the biofilm.
Validation of a new method to isolate single biofilm-embedded cells for persister cell studies.
(A) CLSM of S. epidermidis RP62A biofilms treated with antibiotics at MBCbiofilm suggested isolation of single biofilm cells is necessary for persister cell quantification. (B) SEM of “single cells” isolated from mature biofilms with (left) or without filtration (right). (C) Quantitative comparison of densities (CFU/mL) of suspensions of single biofilm cells before and after filtration. (D) FACS of single biofilm cells before and after filtration.
Population analysis of S. epidermidis biofilm cells
In contrast to the conventional notion that two subpopulations, antibiotic-susceptible bacteria and persister cells comprise a biofilm8,15,44, S. epidermidis biofilm cells presented three subpopulations in response to ciprofloxacin challenge (Fig. 2A). Most cells (>99.99%) were susceptible to ciprofloxacin at concentrations of 0.25–1 mg/L. The remaining cells, namely persister cells (~0.01% for the laboratory reference strain S. epidermidis RP62A and ~0.002% for a S. epidermidis clinical strain isolate 3), were tolerant of ciprofloxacin at concentrations ranging from 1 to 128 mg/L; most of these persister cells (~96.9% of RP62A persister cells and ~96.3% for isolate 3) could be further eradicated by ciprofloxacin at concentrations of 256–1024 mg/L. The third fraction of cells, dormant cells (~3.1% of RP62A persister cells and ~3.7% of isolate 3 persister cells), survived ciprofloxacin at a concentration as high as 1024 mg/L. Typical two-subpopulation patterns were established when vancomycin was used in place of ciprofloxacin. Over 99.9% of biofilm cells were susceptible, responding to vancomycin at concentrations below 4 mg/L. Persister cells (~0.06% for RP62A and 0.03% for isolate 3) demonstrated tolerance to vancomycin at concentrations ranging from 4 to 2048 mg/L (for RP62A) or 128 mg/L (for isolate 3). However, the two-subpopulation pattern could be extended as a three-subpopulation pattern when the incubation period was continued for vancomycin at MBCbiofilm for another 24 h (Fig. 2B). Upon 48 h exposure, only very few dormant cells from the persister cell population, approximately 1.8% for RP62A and 4.6% for isolate 3, survived vancomycin challenge. A similar pattern was found when oxacillin was used to select biofilm persister cells (data not shown). Thus we chose 24 h exposure for ciprofloxacin, or 48 h for vancomycin or oxacillin, at a concentration of MBCbiofilm (Table 1) to select for biofilm dormant cells.
Population analysis of single biofilm-embedded cells.
Population analysis profiling of single S. epidermidis biofilm cells with antibiotics at increasing concentrations identified three fractions of cells of different antibiotic tolerance. The first fraction represents the sensitive subpopulation; the majority of the population were killed following exposure to antibiotics at concentrations close to MBCs (4–8 mg/L for vancomycin and 0.5–1 mg/L for ciprofloxacin). The second fraction represents the persister cell population, which includes both TBK cells and dormant cells. This is a small population remaining tolerant to antibiotics at concentrations ranging from MBCs to sub-MBCbiofilm. The third fraction became visible on further increasing the antibiotic concentration (for ciprofloxacin) and treatment period (for vancomycin) to eradicateTBK cells and to select dormant cells.
A biofilm population contains more tolerant-but-killable cells and dormant cells than a log-planktonic culture
Dormant cells were not detected after exposure of log-planktonic cell cultures to any of the three antibiotics at the levels of MBCbiofilm for 24 or 48 h [limit of detection 10 colony forming units (CFU)/mL], however a substantial number of dormant cells (approximately 1 out of 106 CFU to 1 out of 104 CFU, corresponding to cell proportions 0.0001% to 0.01%) remained viable after exposure of single biofilm cells to these antibiotics (Fig. 3A,B, right panels). Fewer dormant cells were detected after exposure of RP62A or isolate 3 biofilm cells to ciprofloxacin than to vancomycin or oxacillin, with average cell proportions reached 0.00025% and 0.0001% for ciprofloxacin, 0.002% (P = 0.01) and 0.0013% (P = 0.004) for vancomycin and 0.013% (P = 0.005) and 0.004% (P < 0.001) for oxacillin respectively (Fig. 3A,B).
Proportions of S. epidermidis persister cells in two different growth modes.
Total persister cells were isolated by exposing cell cultures to antibiotics at 8 X MIC for 3 h. Dormant cells were selected by exposing cell cultures to antibiotics at MBCbiofilm for 24 or 48 h. The proportion of TBK cells was calculated as the proportion of persister cells minus the proportion of dormant cells. Shown are the averages of three biological repeats in technical triplicates and the standard errors.
TBK cells were isolated from both log-planktonic cultures and biofilms. Cell proportions of TBK cells ranged from 46.7% to 0.023% for individual isolates, different growth mode and antibiotics (Fig. 3A,B, left panels). For both S. epidermidis RP62A and isolate 3, biofilm cell populations generally produced a higher proportion of TBK cells than log-planktonic growth modes (P < 0.05) (Fig. 3A,B, left panels).
Dormant cells are transiently resistant to antibiotics
It has been widely reported that generally defined “persister cells” are only transiently resistant to antibiotics. To determine whether the more stringently selected dormant cells were not genetically resistant to antibiotics, resuscitated dormant cells isolated from a S. epidermidis RP62A biofilm were examined for antibiotic susceptibilities using standard micro-dilution method. Regardless of the method used to isolate dormant cells, the MICs were not different from those of their parent cells (Table 2).
SEM of TBK cells and dormant cells isolated from a S. epidermidis RP62A biofilm
Biofilm persister cells selected with either ciprofloxacin (3 h, 8 X MICs) or oxacillin (24 h, MBCbiofilm) and comprising mostly TBK showed normal cell morphology when assessed by scanning electron microscopy (SEM, Fig. 4A,B). Some cells were in the process of cell division, indicating the presence of metabolically active dividing bacteria (Fig. 4A,B). In contrast, no dividing cells were found in the more stringently selected dormant cell population isolated using vancomycin and oxacillin at MBCbiofilm for 48 h. These cells appeared to have a smaller size, but with a normal shape (Fig. 4C,D).
SEM of TBK cells and dormant cells isolated from S. epidermidis RP62A biofilms.
(A) TBK cells isolated with ciprofloxacin (3 h, 8 x MIC). (B) TBK cells isolated with oxacillin (24 h, MBCbiofilm). (C) Dormant cells isolated with vancomycin (48 h, MBCbiofilm). (D) Dormant cells isolated with oxacillin (48 h, MBCbiofilm). Scale bar = 5 μm.
Antibiotic regimens targeting dormant cells are required to eradicate S. epidermidis biofilms
We hypothesized that choosing antibiotic regimens to target dormant cells might lead to eradication of S. epidermidis biofilms. These regimens can be achieved by using effective antibiotics at adequate concentrations for an extended period, as population analysis suggested the proportion of persister cell is related to the selecting antibiotic, its concentration and treatment period (Fig. 2). We examined the efficacy of ciprofloxacin and vancomycin, as single agents or in simultaneous combination for 48 h in completely killing persister cells and intact biofilms. At MBCbiofilm, ciprofloxacin alone or combinations of ciprofloxacin and vancomycin, but not vancomycin alone, killed nearly all persister cells or biofilm cells of S. epidermidis isolate 3 (see signs in bold, Table 3). Survivors were found for S. epidermidis RP62A, which forms biofilms with more biomass than isolate 3 (data not shown). When 72 h and sequential combinations replaced 48 h and simultaneous combinations, ciprofloxacin or its combination with vancomycin at MBCbiofilm as the primary agent killed nearly all persister cells or biofilm-embedded cells of both isolates (in bold, Table 4). Sequential combinations with ciprofloxacin at MBCbiofilm as the primary agent for the first 24 h, followed by vancomycin at serum-achievable concentration (Table 1) for another 48 h on one occasion eradicated the persister cells and biofilms of isolate 3 (Table 4). In contrast, when antibiotics were used at serum-achievable concentrations, none of the treatments eradicated either persister cells or S. epidermidis biofilms, leaving a substantial number of survivor cells (Tables 3 and 4). To be noted, intact biofilms were occasionally more tolerant to antibiotic treatments than isolated persister cells. For instance, sequential combinations with ciprofloxacin (MBCbiofilm) as the first agent followed by vancomycin (MBCbiofilm) as the second agent killed almost all persister cells but left a substantial number of survivor cells within the biofilms.
Discussion
Highlights of the current study include 1) development of a new method to recover single S. epidermidis biofilm-embedded cells free of biofilm-related antibiotic-compromising factors, 2) demonstrating the presence of three subpopulations of biofilm cells with regard to antibiotic susceptibility and 3) suggesting strategies using antibiotic regimens to eradicate S. epidermidis biofilms.
To our knowledge, this is the first study that successfully isolated large numbers of single cells from bacterial biofilms to study their persister cell proportions. A combination of scraping, pipetting, sonication and vortexing was used to dissociate biofilm cells from the extracellular matrix, followed by a filtration step to select the single cells. The method was qualitatively and quantitatively validated by SEM, fluorescence-activated cell sorting (FACS) and viable counts before and after filtration. Antibiotic-compromising effects of some other biofilm-associated factors, such as extracellular hydrolytic enzymes and QS factors27,28,29,30,31, though not specifically examined, were believed to be minimized by resuspending the biofilm cell into fresh medium after removing from the substratum and before sonication, voxtexing and filtration.
Using the successfully isolated single biofilm cell population, this study provides quantitative and qualitative insights into the presence of antibiotic-tolerant cells within S. epidermidis biofilms. Population analysis suggested that biofilms contain TBK cell and dormant cell subpopulations in addition to normal cells. We showed that dormant cells constitute a very small subpopulation of cells that show little susceptibility to antibiotics, irrespective of antibiotic concentration. These cells were isolated under very stringent conditions that eradicated TBK cells. The rationality of using these stringent conditions is supported by our SEM findings. Both replicating and non-replicating persister cells were found when isolation was carried out following the method published by others6,8,41; however only non-dividing dormant cells were observed when specific concentrations and exposure times based on a pre-determined population analysis profile were introduced (Figs 2 and 4). TBK cells represent a much larger subpopulation than dormant cells. These cells have reduced antibiotic susceptibilities but remain eradicable with selected antibiotics. Viable but non-cultivable cells constitute the fourth cell population that might reside in biofilms14, however, this subpopulation is out of the scope of this study.
Our population profiling of biofilm persister cells based on antibiotic tolerance matches the persister classification by Babalan et al. and others7,10,42,45. Babalan et al. categorized persister cells as either type I or type II10. Type I persister cells are a fully dormant sub-population that can only be isolated from stationary-phase cultures; type II persister cells are a fraction of cells in a slow-growth state that can be obtained from any stages of growth, including log-planktonic cultures10,45. Dormant cells described in the present study are similar to type I persister cells. These cells were mainly isolated from the biofilm, a growth mode that has similar levels of resistance to antibiotics as stationary-phase cultures; SEM images of dormant cells showed no evidence of cell division. TBK cells might characteristically match the continuously generated type II persister cells. In our study, TBK cells could be isolated from both biofilms and log-planktonic cultures and SEM clearly showed dividing cells. Our finding that a single cell population isolated from S. epidermidis biofilms contained more persister cells than log-planktonic cultures is consistent with other published studies3,4,39, however this study goes into greater detail than previous studies. The observation presented in this study that dormant cells are only found in the biofilm growth mode suggests their important role in the tolerance of biofilms to high concentrations of antibiotics.
Zhang et al. has suggested a key principal for successful treatment of chronic infections: to reduce the persister cells quickly to a sufficiently low number with effective drugs so that reversion from persister cells to a normal bacterial population will not occur before the treatment is continued46,47. By introducing this principal into our study, we proposed a new strategy to treat S. epidermidis biofilm: killing TBK cells with effective antibiotics within a short period, followed immediately by a dormant-cell-targeted antibiotic regimen. Our population profiling suggested that high doses of ciprofloxacin but not vancomycin or oxacillin eradicate S. epidermidis TBK cells within 24 h. This is in accordance with the high efficacy of ciprofloxacin against staphylococcal biofilms48,49. We also observed that the critical concentration, MBCbiofilm must be reached to completely kill the TBK and to expose dormant cells. To further eradicate dormant cells, we propose extending the treatment period to a continuous 48 h or 72 h (Tables 3 and 4). In our in vitro study, eradication of dormant cells and intact biofilms were both achieved with ciprofloxacin at MBCbiofilm, or on some occasions, combinations with ciprofloxacin at MBCbiofilm as the major agent. Extended incubation might “wake up” the persister cells from dormancy and recover their susceptibility to antibiotics before they can establish a new population50,51. We however cannot exclude the possibility that the killing of dormant cells is simply a consequence of a longer exposure to antibiotics.
We initially proposed that combining vancomycin and ciprofloxacin would be more efficient in killing persister cells than ciprofloxacin alone, based on the staggered tolerance spectrum of persister cells to vancomycin and ciprofloxacin and also on conclusions from other studies8. Surprisingly, combinations of vancomycin and ciprofloxacin at MBCbiofilm generally demonstrated a lower efficacy in eradicating persister cells or intact biofilms than ciprofloxacin alone. This might be explained by the finding by Dorr et al. that stress from antibiotics can stimulate the formation of persister cells52. To be noted, there is also a slight difference in antibiotic tolerance between single biofilm cell populations and intact biofilms (Tables 3 and 4), suggesting the possible involvement of other biofilm-related factors, such as the presence of EPS, QS factors and extracellular hydrolytic enzymes.
In summary, S. epidermidis biofilm persister cells comprise TBK cells and dormant cells; both subpopulations play important roles in biofilm drug tolerance. Selection of a suitable antibiotic, at adequate concentration and an extended exposure time, should be considered in the treatment of S. epidermidis biofilm-related infections. Although the concentrations of antibiotics and treatment period used in this in vitro study might not be suitable for intravenous administration, they could be applied for other applications such as antibiotic lock solutions, inhaled aerosolized antibiotics or wound dressings53,54.
Materials and Methods
Bacterial strains and antibiotics
Two biofilm-positive S. epidermidis strains: RP62A (ATCC 35984), resistant to oxacillin but sensitive to vancomycin; and isolate 3, sensitive to oxacillin and vancomycin, were used in this study. S. epidermidis isolate 3 is a clinical isolate, originally obtained from a blood culture from a baby being cared for in the Neonatal Intensive Care Unit, Royal Women’s Hospital, Melbourne55. First line antibiotics, oxacillin, vancomycin and ciprofloxacin used in this study were purchased from Sigma (Sigma–Aldrich, Sydney, Australia).
Biofilm assays and isolation of single biofilm cells
In order to isolate single cells from S. epidermidis biofilms, we combined a filtration step with mechanical disruption to separate biofilm cells from biofilm-associated factors that might impact on antibiotic efficacy. Staphylococcus epidermidis biofilms were grown in 6-well microplates with Tryptone Soya Broth (TSB, Oxoid, Hampshire, England) as described by Deighton et al.56. Single biofilm cells were isolated as follows: after preparing and washing biofilms, one milliliter of Muller-Hinton broth (MHB, Oxoid, Hampshire, England) was added to each well and the wells were scraped thoroughly with a FALCON® cell scraper (Corning, Mexico). The resulting suspensions were mixed well by pipetting several times and transferred into a sterile tube, followed by vigorous vortexing. This step was repeated three times and the final volume of each suspension was adjusted to 25 mL with MHB. The tube was sonicated for 10 min using a sonication bath (42 KHZ, BRANSON 1510) and then vortexed at the highest speed for 2 min (30″ × 4) to break up cell aggregates. The suspensions were firmly pressed through 1.2 μm Acrodisc syringe filters. Muller-Hinton broth was chosen as it is recommended by Clinical Laboratory and Standard Institute (CLSI) for antibiotic susceptibility testing and has been used previously in persister cell studies23.
Preparation of single planktonic cells at mid-log phase
Planktonic cultures at mid-log phase were prepared by inoculating one colony of overnight-grown S. epidermidis into 5 mL nutrient broth (NB, Oxoid, Hampshire, England). Cultures were grown at 37 °C for 4–5 h until the turbidity was equivalent to McFarland 0.5 standard (~1 × 108 CFU/mL). Bacterial suspensions were then centrifuged at 4400 g for 5 minutes, washed twice with sterile phosphate buffered saline (PBS), re-suspended in MHB, followed by the same vortex and filtration procedures described for biofilm cells.
Population analysis of biofilm embedded cells
A published method for population analysis profiling was modified for this study as persister cells are unable to pass their “resistant” trait to descendants under a stress-free condition and cannot re-grow into a new population such as a colony on agar plates containing antibiotics51,57. To profile the population of S. epidermidis RP62A and isolate 3 biofilm cells upon antibiotic exposure, suspensions of single cells at ~107 CFU/mL were prepared with MHB containing vancomycin or ciprofloxacin at increasing concentrations ranging from planktonic MBCs to MBCbiofilm (see below). Viable counts were performed after exposure of biofilm cells to vancomycin and ciprofloxacin for 24 h respectively, or exposure to vancomycin at MBCbiofilm (2048 mg/L) for 48 h. The rationale of selecting such concentrations of antibiotics was to ensure the effective killing of antibiotic-sensitive cells and TBK cells in the bacterial populations. MBCs of antibiotics for planktonic cells at mid-log phase were determined by the broth micro-dilution method recommended by CLSI58. MBCbiofilm of oxacillin, vancomycin and ciprofloxacin for biofilms formed by S. epidermidis RP62a and isolate 3 were determined in our previous study (Table 1)35. To avoid antibiotic carryover which is related to the usage of very high concentration of antibiotics that might hinder the recovery of viable cells on agar plates, bacterial suspensions were pelleted by centrifuging at 20,000 g for 5 minutes, washed three times with PBS before being plated on Typtone Soya Agar (TSA) plates. Colonies were counted after 72 h of incubation at 37 °C. The log10 of CFU/mL was plotted against the antibiotic concentration. The difference in the number of survivors after exposure of cells to two antibiotic concentrations represents the fraction of cells that remains sensitive to the higher concentration but tolerant to the lower concentration of antibiotics.
Quantification of tolerant-but-killable cells and dormant cells
We also used a previously reported method to quantify the generally defined persister cells from planktonic cultures at mid-log phase and single biofilm cell populations6,41. To isolate the generally defined “persister cell” population, which might comprise both TBK cells and dormant cells, suspensions of single cells at densities of ~108 CFU/mL from planktonic cultures at mid-log phase and biofilms were exposed to antibiotics at 8 × MIC for 3 h6,41. This method has been shown to eradicate the majority of bacteria with normal susceptibilities8. Eight times of MIC of the selected antibiotics are slightly higher than their MBCs, but remain achievable in human serum (Table 1). Before and after the antibiotic challenge, 100 μL volumes of suspension were sampled, serially diluted with PBS and 10 μL was spotted on a TSA plate for cell counting. In some cases 100 μL volumes of the suspension were directly spread on agar plates to increase the sensitivity for detecting persister cells. Cultures were incubated for 72 h to maximize recovery of persister cells.
To isolate dormant cells, suspensions of single cells at mid-log phase or single biofilm cells were statically exposed to antibiotics at MBCbiofilm for the designated exposure time (37 °C, 24 h for ciprofloxacin and 48 h for vancomycin or oxacillin, based on our population analysis results). The same treatment periods were used for both log-planktonic cultures and biofilm cells to enable valid comparisons to be made. The numbers of survivors were determined by viable count as described above. The proportion of persister cells and dormant cells in different bacterial populations was calculated as follows: (bacterial density after antibiotic treatment)/(bacterial density before antibiotic treatment). The proportion of TBK cells was calculated as the proportion of persister cells minus the proportion of dormant cells.
Antibiotic susceptibilities of dormant cells
In order to determine whether there was a resistance determinant passed from parent cells to the strictly selected dormant cells, antibiotic susceptibility tests were performed for descendants of dormant cells. Three to five colonies from the dormant cell quantification procedure (grown on TSA plate after antibiotic treatment) were inoculated directly into MHB and grown for 5 h. The resulting suspensions were used for the MIC tests, following the CLSI guideline.
Scanning electron microscopy
Scanning electron microscopy was employed to examine the presence/absence of bacterial cell aggregations before and after filtration when isolating single cells from biofilms and also to examine the morphology of biofilm persister cells and dormant cells. After isolating persister cells with ciprofloxacin (8 X MIC for 3 h), oxacillin (8 X MIC for 24 h) and dormant cells with oxacillin (MBCbiofilm for 48 h) and vancomycin (MBCbiofilm for 48 h) respectively, 100 μL of the suspensions were placed on filter membranes (0.2 μm or 0.45 μm) and air-dried overnight. The samples were sputter coated with 200 angstroms of gold using a Magnetron sputter coater (Dynavac, SC100M) and the trapped cells were viewed and imaged with scanning electron microscopes (Philips XL30 or Hitachi S570).
Eradication of persister cells or intact biofilms with antibiotics
Intact S. epidermidis biofilms were formed in 96-well microplate56 and single biofilm cells were prepared as described above. Both biofilms and single biofilm cell populations were challenged with vancomycin and ciprofloxacin for 48 h, alone or in simultaneous combinations at MBCbiofilm or serum achievable concentration (see Table 3). In parallel, we challenged intact biofilms or isolated single biofilm cells with single agent for 72 h or two antibiotics sequentially for a total exposure period of 72 h. Two protocols were examined: vancomycin for 48 h and then ciprofloxacin for 24 h, or vice versa (Table 4). Viable counts were carried out for survivor persister cells as described earlier. To increase the detection sensitivity, a one mL volume of the treated suspension was pelleted, washed three times with PBS and resuspended into 100 uL of PBS, followed by spreading and recovering on TSA plates. To perform viable count for survivors from intact biofilms, biofilms cells were scraped and removed from the bottom of a 96-well microplate and transferred in an Eppendorf tube. After dissociating the cell clumps by vortex and washing with PBS three times, the cells were plated on TSA plate and grown at 37 °C for three days.
Additional Information
How to cite this article: Yang, S. et al. Antibiotic regimen based on population analysis of residing persister cells eradicates Staphylococcus epidermidis biofilms. Sci. Rep. 5, 18578; doi: 10.1038/srep18578 (2015).
References
Percival, S. L., Hill, K. E., Malic, S., Thomas, D. W. & Williams, D. W. Antimicrobial tolerance and the significance of persister cells in recalcitrant chronic wound biofilms. Wound Repair Regen. 19, 1–9 (2011).
Roberts, M. E. & Stewart, P. S. Modelling protection from antimicrobial agents in biofilms through the formation of persister cells. Microbiology 151, 75–80 (2005).
Lewis, K. Persister cells and the riddle of biofilm survival. Biochemistry (Mosc) 70, 267–274 (2005).
Spoering, A. L. & Lewis, K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183, 6746–6751 (2001).
Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56 (2007).
Keren, I., Kaldalu, N., Spoering, A., Wang, Y. & Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13–18 (2004).
Luidalepp, H., Joers, A., Kaldalu, N. & Tenson, T. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J. Bacteriol. 193, 3598–3605 (2011).
Lechner, S., Lewis, K. & Bertram, R. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J. Mol. Microbiol. Biotechnol. 22, 235–244 (2012).
Lewis, K. Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Lechner, S., Patra, P., Klumpp, S. & Bertram, R. Interplay between population dynamics and drug tolerance of Staphylococcus aureus persister cells. J. Mol. Microbiol. Biotechnol. 22, 381–391 (2012).
Shah, D. et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6, 53 (2006).
Keren, I., Minami, S., Rubin, E. & Lewis, K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2, e00100–00111 (2011).
Orman, M. A. & Brynildsen, M. P. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob. Agents Chemother. 57, 3230–3239 (2013).
Kint, C. I., Verstraeten, N., Fauvart, M. & Michiels, J. New-found fundamentals of bacterial persistence. Trends Microbiol. 20, 577–585 (2012).
Barth, Jr. V. C. et al. Heterogeneous persister cells formation in Acinetobacter baumannii. PLoS One 8, e84361 (2013).
Davey, M. E. & O’Toole G., A. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Bio. Rev. 64, 847–867 (2000).
Costerton, J. W., Stewart, P. S. & Greenberg, E. P. Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 (1999).
Hall-Stoodley, L. & Stoodley, P. Evolving concepts in biofilm infections. Cell Microbiol. 11, 1034–1043 (2009).
Aaron, S. D. et al. Single and combination antibiotic susceptibilities of planktonic, adherent and biofilm-grown Pseudomonas aeruginosa isolates cultured from sputa of adults with cystic fibrosis. J. Clin. Microbiol. 40, 4172–4179 (2002).
Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 34, 877–886 (2015).
Mah, T. F. Biofilm-specific antibiotic resistance. Future Microbiol. 7, 1061–1072 (2012).
Mulcahy, L. R., Burns, J. L., Lory, S. & Lewis, K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192, 6191–6199 (2010).
Muranaka, L. S., Takita, M. A., Olivato, J. C., Kishi, L. T. & de Souza, A. A. Global expression profile of biofilm resistance to antimicrobial compounds in the plant-pathogenic bacterium Xylella fastidiosa reveals evidence of persister cells. J. Bacteriol. 194, 4561–4569 (2012).
Barraud, N., Buson, A., Jarolimek, W. & Rice, S. A. Mannitol enhances antibiotic sensitivity of persister bacteria in Pseudomonas aeruginosa biofilms. PLoS One 8, e84220 (2013).
Wang, X. & Wood, T. K. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77, 5577–5583 (2011).
Khakimova, M., Ahlgren, H. G., Harrison, J. J., English, A. M. & Nguyen, D. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J. Bacteriol. 195, 2011–2020 (2013).
Dorr, T., Lewis, K. & Vulic, M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 5, e1000760 (2009).
De Leenheer, P. & Cogan, N. G. Failure of antibiotic treatment in microbial populations. J. Math. Biol. 59, 563–579 (2009).
Stewart, P. S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292, 107–113 (2002).
Mah, T. F. & O’Toole, G. A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39 (2001).
Lewis, K. Persister Cells. Annu. Rev. Microbiol. (2010).
Shapiro, J. A., Nguyen, V. L. & Chamberlain, N. R. Evidence for persisters in Staphylococcus epidermidis RP62a planktonic cultures and biofilms. J. Med. Microbiol. 60, 950–960 (2011).
Lewis, K. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322, 107–131 (2008).
Qu, Y., Daley, A. J., Istivan, T. S., Rouch, D. A. & Deighton, M. A. Densely adherent growth mode, rather than extracellular polymer substance matrix build-up ability, contributes to high resistance of Staphylococcus epidermidis biofilms to antibiotics. J. Antimicrob. Chemother. 65, 1405–1411 (2010).
Brooun, A., Liu, S. & Lewis, K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44, 640–646 (2000).
LaFleur, M. D., Kumamoto, C. A. & Lewis, K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 50, 3839–3846 (2006).
Al-Dhaheri, R. S. & Douglas, L. J. Absence of amphotericin B-tolerant persister cells in biofilms of some Candida species. Antimicrob. Agents Chemother. 52, 1884–1887 (2008).
Singh, R., Ray, P., Das, A. & Sharma, M. Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: an in vitro study. J. Med. Microbial. 58, 1067–1073 (2009).
Chen, X., Zhang, M., Zhou, C., Kallenbach, N. R. & Ren, D. Control of bacterial persister cells by Trp/Arg-containing antimicrobial peptides. Appl. Environ. Microbiol. 77, 4878–4885 (2011).
Keren, I., Shah, D., Spoering, A., Kaldalu, N. & Lewis, K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186, 8172–8180 (2004).
Ma, C. et al. Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol. Lett. 303, 33–40 (2010).
Li, Y. & Zhang, Y. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51, 2092–2099 (2007).
Lewis, K. Persister cells: molecular mechanisms related to antibiotic tolerance. Handb. Exp. Pharmacol. 211, 121–133 (2012).
Canas-Duarte, S. J., Restrepo, S. & Pedraza, J. M. Novel protocol for persister cells isolation. PLoS One 9, e88660 (2014).
Byrne, S. T. et al. Pyrrolidine dithiocarbamate and diethyldithiocarbamate are active against growing and nongrowing persister Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 51, 4495–4497 (2007).
Zhang, Y. Advances in the treatment of tuberculosis. Clin. Pharmacol. Ther. 82, 595–600 (2007).
Knobloch, J. K., Von Osten, H., Horstkotte, M. A., Rohde, H. & Mack, D. Biofilm formation is not necessary for development of quinolone-resistant “persister” cells in an attached Staphylococcus epidermidis population. Int. J. Artif. Organs 31, 752–760 (2008).
Knudsen, G. M., Ng, Y. & Gram, L. Survival of bactericidal antibiotic treatment by a persister subpopulation of Listeria monocytogenes. Appl. Environ. Microbiol. 79, 7390–7397 (2013).
Joers, A., Kaldalu, N. & Tenson, T. The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. J. Bacteriol. 192, 3379–3384 (2010).
Wood, T. K., Knabel, S. J. & Kwan, B. W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 79, 7116–7121 (2013).
Dorr, T., Vulic, M. & Lewis, K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8, e1000317 (2010).
Ballmann, M., Smyth, A. & Geller, D. E. Therapeutic approaches to chronic cystic fibrosis respiratory infections with available, emerging aerosolized antibiotics. Respir. Med. 105 Suppl 2, S2–8 (2011).
Bookstaver, P. B., Gerrald, K. R. & Moran, R. R. Clinical outcomes of antimicrobial lock solutions used in a treatment modality: a retrospective case series analysis. Clin. Pharmacol. 2, 123–130 (2010).
Bradford, R. et al. Coagulase-negative staphylococci in very-low-birth-weight infants: inability of genetic markers to distinguish invasive strains from blood culture contaminants. Eur. J. Clin. Microbiol. Infect. Dis. 25, 283–290 (2006).
Deighton, M. A., Capstick, J., Domalewski, E. & Van Nguyen, T. Methods for studying biofilms produced by Staphylococcus epidermidis. Methods Enzymol. 336, 177–195 (2001).
Wootton, M. et al. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47, 399–403 (2001).
CLSI CLSI, Clinical and Laboratory Standards Institute. M7-A7, Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard (7th ed.) Wayne, PA. (Wayne, PA; 2004).
Acknowledgements
We thank Mrs Joan Clark from Monash Micro Imaging for her assistance with scanning electron microscopy. We thank Monash University (Interdisciplinary Research Fund seeding grant to Y.Q.), School of Applied Sciences, RMIT University and Australian Research Council (ARC) for the funding. Y.Q. is an ARC Super Science Fellow.
Author information
Authors and Affiliations
Contributions
Y.Q., M.A.D. and S.Y. designed the project. S.Y., Y.Q., I.D.H. and M.S. performed the experiments. S.Y., Y.Q., D.R.C., B.C., F.S., A.Y.P. and T.L. interpreted data. Y.Q. and M.A.D. wrote the main manuscript text. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yang, S., Hay, I., Cameron, D. et al. Antibiotic regimen based on population analysis of residing persister cells eradicates Staphylococcus epidermidis biofilms. Sci Rep 5, 18578 (2016). https://doi.org/10.1038/srep18578
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep18578
This article is cited by
-
Antibiofilm activity of marine microbial natural products: potential peptide- and polyketide-derived molecules from marine microbes toward targeting biofilm-forming pathogens
Journal of Natural Medicines (2024)
-
Multidrug efflux pumps: structure, function and regulation
Nature Reviews Microbiology (2018)
-
How do environment-dependent switching rates between susceptible and persister cells affect the dynamics of biofilms faced with antibiotics?
npj Biofilms and Microbiomes (2018)
-
Efflux drug transporters at the forefront of antimicrobial resistance
European Biophysics Journal (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.