Abstract
Enteroviruses are known to cause childhood paralysis. The purpose of this study was to examine the genetic diversity and to determine the association of non-polio enteroviruses (NPEVs) with acute flaccid Paralysis (AFP). Stool samples (n = 1191) of children with AFP were collected from Khyber Pakhtunkhwa and Federally Administered Tribal Areas of Pakistan. Poliovirus was isolated in 205 (17.2%) samples and NPEVs were found in 215 (18.0%) samples. Out of 215 viruses, 124 (57.7%) were typed into 19 different types of enteroviruses while 91 (42.3%) remained untypeable on microneutralization assay that were reconfirmed as NPEVs by real time PCR. Echovirus 19 (20/35; 57.1%) was found the most prevalent type based on VP1 nucleotide sequencing with increased genetic diversity. Phylogenetic analysis revealed the circulation of a new genotype of E-19 in the country. The findings of this study are of great importance for future research and propose to establish the enterovirus surveillance system in the country to readily identify more enteroviruses and to monitor the emergence of new variants/genotypes especially at the moment when we are at the verge of polio eradication phase.
Similar content being viewed by others
Introduction
Enteroviruses (EVs) are small (22 to 30 nm) non-enveloped, single-stranded RNA viruses comprising four viral capsid proteins VP1–41. Traditionally, enteroviruses are classified into four groups (i) polioviruses (ii) coxsackieviruses A (iii) coxsackieviruses B (iv) enteric cytopathogenic human orphan (ECHO) viruses based on pathogenesis of infection in humans and experimental suckling mice. Later on, neutralization with type specific antisera was used for classification of these viruses2,3 but this approach failed to provide consistent results due to low sensitivity of virus isolation and emergence of many untypable/new types4,5. As a result, a molecular approach targeting VP1 encoding region was introduced for characterization of enteroviruses and it was proposed that EVs shall be classified as the same type if they share >75% nucleotide (>85% amino acid) identity in the VP1 coding sequence and into a new type if the nucleotide similarity is <75% with existing types1,6,7.
However, at present, based on the phylogenetic relationships in multiple genome regions, International Committee on Taxonomy of Viruses (ICTV) reclassified enteroviruses into twelve species, i.e., EV-A to H, EV-J and human rhinovirus (HRV)-A to C8,9,10. Only seven species (EV-A to D and HRV-A to C) infect humans while remaining infect cattle (bovine enteroviruses, porcine enteroviruses), ape species and monkey (simian enteroviruses). The EV-B is most diverse species with at least 61 types, including coxsackievirus (Cox) B1-6, A9, Echovirus (E) E1–7, 9, 11–21, 24–27, 29–33 and EV-B 69, 73–75, 77–88, 93, 97–98, 100–101, 106–107, 110–111 and Simian agent 5 (SA5), which are closely related to simian enteroviruses11,12.
Most enteroviral infections are asymptomatic; however, sometimes they cause mild to severe illness such as aseptic meningitis, encephalitis conjunctivitis, myocarditis, poliomyelitis, neonatal enteroviral sepsis and acute flaccid paralysis13,14.
Echovirus 19 was first isolated from stool sample of a child suffering from diarrhea in 195515 and later from cerebrospinal fluid (CSF) of a man with aseptic meningitis in 195916. However, epidemic of E-19 infection among infants and children has been observed in Europe during 1974 and 197517,18. Similarly, association of this virus with other infections such as epidemic neuromyasthenia, uveitis, gastroenteritis and myalgia has been reported in previous years19 and recently echovirus 19 has been isolated from a case of AFP child in Australia20.
In this study, we analyzed stool samples collected from acute flaccid paralytic children residing in Khyber Pakhtunkhwa (KP) and Federally Administered Tribal Areas (FATA) of Pakistan to investigate the association of NPEVs with AFP and to examine the genetic diversity among these viruses. The genetic data reveals that echovirus 19 is the most prevalent serotype and has some possible link in causation of paralysis. Furthermore, genome analysis confirms the circulation of a new indigenous genotype of E-19 with increased genetic diversity in the country.
Results
Enterovirus Cell Culture and Microneutralization
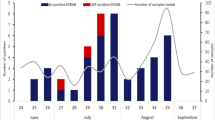
A total of 1191 stool samples (887 from Khyber Pakhtunkhwa and 304 from FATA; each represents a unique patient) collected during January to December 2013 from AFP patients were processed and inoculated on RD and L20B cell lines. Poliovirus was detected in 205 (17.2%) samples and non polio enteroviruses were found in 215 (18.0%) samples (Table 1). Out of 215 samples (n = 45 from FATA and n = 170 from KP) 124 (57.7%) samples were successfully typed into 19 different enterovirus serotypes (E-1-7, 11-14, 20, 21, 25, 27, 29, 30, 33 and CV-A9) while 91 (42.3%) remained untypeable by microneutralization assay.
Molecular Typing
All untypeable isolates (n = 91) were reconfirmed as NPEVs by real-time reverse transcriptase polymerase chain reaction targeting 5′-UTR21 and nucleotide sequencing of a partial VP1 specific fragment22 placed 97.8% of these isolates into EV-B and 2.2% into EV-C species (data not shown). It has been found that majority (35/89; 39.3%) were echoviruses with more prevalent type Echovirus 19 (20/35; 57.1%). The clinical features of patients having AFP associated with E-19 are shown in Table 2.
VP1 sequence analysis of Pakistan Echovirus 19 isolates
To investigate the molecular epidemiology and genetic diversity of most prevalent serotype E-19, partial VP1 nucleotide sequences (399 base pairs) of all 20 isolates were compared with prototype sequences of different enteroviruses retrieved from GenBank. All study strains (n = 20) were found closely related with echovirus 19 prototype (Burke; Accession No. AF081332) having mean 73.7% (73.4–76.9%) nucleotide identity within enterovirus B species and fulfills the new genotype demarcation criterion as described earlier5,23,24,25,26,27 that strain sharing <85% nucleotide identity in VP1 region with prototype sequence should be placed in different genotype. Therefore, it is proposed that Pakistan (PAK) isolates form a new genotype of E-19.
Genetic Diversity among Pakistan Echovirus 19 isolates
Phylogenetic trees were constructed by aligning the partial VP1 sequences of E-19 study strains with their closely related sequences retrieved from GenBank to study their genetic diversity and possible relationships. Pair wise distance scores confirmed that all study isolates constitute a separate clade (bootstrap 85%) that further segregated into two distinct genetic groups (I and II) with 93.3% (89.7–94.8%) mean nucleotide identity with each other supported by high bootstrap value 90 and 86 respectively (Fig. 1a).
(a) A Phylogenetic tree was constructed from the partial VP1 nucleotide sequences (399 nucleotides) of twenty echovirus 19 strains and their closely related sequences retrieved from GenBank (accession numbers are included in the virus names) using maximum likelihood method with 1000 bootstrap by MEGA 5.0. Bootstrap values greater than 50 are indicated at the respective nodes and the scale bar represents the evolutionary distance. Genotype and groups are marked on the tree. The isolates from this study, closely related and prototype strains are represented by ‘●’, ‘♦’ and ‘▲’ taxon markers respectively. (b) Maximum clade credibility (MCC) tree based on the Bayesian analysis of the partial VP1 nucleotide sequences of PAK E-19 and their closely related sequences retrieved from GenBank. The tree was estimated with Hasegawa-Kishino-Yano nucleotide substitution model under the uncorrelated relaxed clock approach (UCLD). Branch tips correspond to date of collection and each tip represents a virus strain in 1 patient. Tree nodes were annotated with posterior probability (pp) values. Horizontal axis indicates time in years. PAK: Pakistan.
Group I comprised 10 isolates from different districts of KP and FATA showing 73.7% (74.0–76.9%) mean nucleotide identity with E-19 prototype strain. All isolates in Group I presented 96.6% (91.7–98.9%) mean nucleotide identity with each other and one isolate PAK-MEV3422B isolated from district Malakand was most divergent isolate showing 95.14% (91.7–97.0%) mean nucleotide identity. Further it was observed that the closely related isolate to group I was PAK/RRL-15-2009 (Pakistan strain; Accession No. GU355715) showing 92.1% (87.9–93.8%) mean nucleotide identity.
Similarly, group II contained 10 isolates presenting 73.8% (72.7–75.1%) mean nucleotide identity with E-19 reference strain and 98% (95.4–99.6%) mean nucleotide identity among each other. The most divergent strain in this group was PAK-MEV4343A isolated from district Mardan with 96.1% (95.4–96.6%) mean nucleotide identity and it was also noticed that closely related strain to group II members was PAK/RRL-15-2009 having 92.4% (91.7–92.9%) mean nucleotide identity.
Phylogenetic Analyses of Pakistan Echovirus 19 Isolates
The molecular evolution of PAK E-19 strains was also investigated by phylogenetic analysis together with closely related E-19 VP1 sequences from the GenBank database (Fig. 1a). The phylogenetic tree revealed that study strains of E-19 represent a clade which is completely different from those detected earlier from China, Australia and Central African Republic. All PAK study isolates had distinct genetic relationship with other E-19 strains and constitute a separate genetic clade representing a new genotype of E-19.
In addition, the partial VP1 gene sequences were also analyzed using a Bayesian method to estimate the phylogenetic tree topology, to investigate the genetic relationship of study isolates sequences and to classify them consistently among the current echovirus 19 strains. The Bayesian phylogram topology (Fig. 1b) was congruent with the topology previously observed in Maximum Likelihood tree (Fig. 1a). Overall, all the sequences of E-19 serotype sampled from 1955 to 2013 were distributed into two major partitions identified by ancestral nodes 1 and 2 (Fig. 1b). First partition contained only three isolates reported from Central African Republic. The second major tree partition was characterized by three different clades (1–3) with posterior probability 1. In this partition, a distinct clade 2 corresponding to Pakistan isolates clearly indicates a strong temporal clustering of sequences with two different groups supported by posterior probability 1 (Fig. 1b). Additionally, the study isolates displayed a close relatedness with an earlier identified E-19 PAK isolate (PAK-RRL-15-2009; Accession No. GU355715).
The most recent common ancestor of all PAK isolates dated back to about seven years ago 2006.3 (95% HPD: 2003.9–2008.4 years). Later on, the study isolates has diverged into two groups through node 6 (pp = 1) whose time was estimated to be 2008.7 (95% HPD: 2006.7–2010.5) years. Furthermore, group I was originated first as compared to Group II and the time origin of isolates into group I was estimated to be 2009.9 (95% HPD: 2008.2–2011.3) years while for Group II was 2010.5 (95% HPD: 2009.11–2011.7) years. The evolutionary timescale analysis of E-19 VP1 sequences revealed that the study strains diverged 58 years ago (1955) from their first isolated prototype (Burke). The tree root was estimated to the year 1949 with a credibility interval ranging between the years 1936.1 and 1955 (Fig. 1b).
Amino acid Sequence Comparison
The deduced amino acid sequences (133 amino acids) of VP1 region of all PAK E-19 strains compared with prototype Burke strain and similarity rate was observed 90.5% (88.5–91.8%). Amino acid alignments revealed that there were 105 (78.9%) conserved and 29 (21.1%) variable amino acid sites (Fig. 2). In addition, three substitutions were found in BC loop region (82 to 92 amino acid residues) between E-19 study strains and Burke prototype strains. Among these substitutions, one was glutamine (Q) to lysine (K) at amino acid 82 except in two isolates (PAK-MEV3422B and PAK-MEV4343A) where glutamine was replaced by Asparagine (N) and Arginine (R) respectively. The second substitution was serine (S) to histidine (H) at amino acid 84 and third was aspartic acid (D) to glutamic acid (E) at amino acid 85. No genotype specific amino acid signatures were observed in the sequence.
Alignment of deduced amino acid sequences of the VP1 polyprotein of echovirus 19 strains with the prototype sequence showing only the variable sites.
The sequences within the rectangle show variable sites within the BC-loop region. Strain designations are on the left hand side of the alignment. Dots (.) represent amino acids identical with respect to prototype strain of echovirus 19 and the amino acid symbols indicate where unrelated amino acids exist with reference to prototype strain. In sequence logo the height of the logo at each site is equal to the total information at that site and the height of each symbol in the logo is proportional to its contribution to the information content.
Discussion
Acute flaccid paralysis (AFP) is a heterogeneous neurologic condition defined by the sudden onset of weakness and floppiness in any part of the body in a child <15 years of age or paralysis in a person of any age in whom poliomyelitis is suspected28.The predominant cause of AFP is infection with poliovirus but a number of non polio enteroviruses including coxsackieviruses, echoviruses and enterovirus 68, 70, 71 have also been reported to cause many neurological disorders such as encephalitis, meningitis, meningoencephalitis and AFP29,30,31,32,33. Similarly, in Pakistan many non polio enteroviruses have been isolated from AFP cases but unfortunately, these cases were never being explored further either by the polio eradication programme or the country health care system. As a result, no clear and detailed information about the epidemiology of NPEVs in the country is available except few reports34,35,36,37. Therefore, present study was designed to genetically characterize the NPEVs isolated from paralytic cases, as well as to understand the role of echovirus 19 in acute flaccid paralysis owing to the fact that Pakistan is at the verge to achieve the goal of polio eradication.
Echoviruses frequently cause aseptic meningitis38 but they may involve in paralysis, encephalitis, neonatal sepsis, exanthema, respiratory disease, myalgia, myocarditis, uveitis and hand-foot and mouth disease19,39. It was observed that proportion of echoviruses isolated in this study was high (73.1%) as compared to the numbered enteroviruses (21.7%) and coxsackieviruses (4.6%). Similarly, among echoviruses, the echovirus 19 was found the most prevalent human enterovirus (12.6%). Although echovirus 19 may cause mild to severe neurological disorders as reported from Malaysia, China, United States of America and Australia20,40,41,42,43 but unfortunately, no clinical information, molecular epidemiology and disease correlation of this serotype with AFP have been reported except two case reports. In first case, E-19 was isolated from CSF sample of a 39 year old immunosuppressed transplant recipient who admitted to the hospital with acute onset of flaccid paralysis of the left leg43 and in second case a three years old Australian child admitted to hospital with a 2-day history of fever to 38.8 °C and a 24-h history of painless inability to move her left upper arm and shoulder. A detailed medical history showed weakness restricted to the left upper limb, with power at the shoulder and elbow. The muscle tone was flaccid, but there was some degree of wrist flexion and extension and preservation of finger movements, with an absence of biceps, triceps and brachioradialis reflexes. Her stool sample was processed for NPEVs and confirmed it as E-1920. On discharge from hospital she had physiotherapy and rehabilitation but had residual weakness of her upper limb after 6 months. This case report supports our current study finding that suggests E-19 may have contribution in paralysis. Although, NPEVs are rarely associated with sporadic cases of AFP but we have isolated twenty E-19 strains from paralytic patients reside in a small geographic area of Pakistan. Clinically, all these patients presented fever, asymmetrical paralysis and rapid progression of paralysis at the time of onset of the disease. Additionally, residual weakness was seen in a marked proportion of AFP cases on 60 days follow up investigation. Consequently, all these findings suggest E-19 as the possible causative agent of paralysis and to the best of our knowledge; this is first report about the epidemiological linkage of E-19 with AFP published from Pakistan so far.
Furthermore, all viruses of this study, isolated from AFP cases grouped together in E-19 cluster and form a distinct genotype having 26.3% nucleotide divergence from their prototype and fulfill the criterion for assessment of new genotype as described previously5,23,24,25,26,27,36,44,45. Moreover, it was perceived by Bayesian evolutionary analysis (Fig. 1b), that the most recent common ancestors for both groups 1 and 2 viruses were dated to 2008.7, indicating that the two groups of new E-19 genotype were circulated for ~4.5 years in the community and were not simply the result of any current transmission. The maximum clade credibility (MCC) tree was also notable for the presence of separate clade containing the study isolates that were shown to have diverged into two groups and been introduced from China in year 2006.2 in the country. Similarly, the frequent presence of long branches in the tree point out towards increased genetic diversity that indicates a gap, probably created by a small number of samples collected and analyzed during the study period.
The BC loop in the VP1 region is, in particular, one of the regions associated with viral antigenicity and substitution mutations may result in changes in this region, can play a role in host adaptation for enteroviruses46. All PAK E-19 isolates had three substitutions within the BC loop on comparison with prototype. Therefore, the effect of these mutations may need further investigations to rule out any changes in antigenic and biochemical properties.
In conclusion, we presented a genetic overview of E-19 with increased genomic diversity and explored its possible association with acute flaccid paralysis. Although Pakistan is a polio endemic country where more efforts are focused on poliovirus eradication but there is a dire need to assess the impact of the disease burden caused by this group of viruses as polio vaccine offers no protection against NPEVs infection. Moreover, we speculate that NPEVs may predominate in its toll to cause paralysis after post-eradication. Therefore, enterovirus surveillance is needed to strengthen in the country in order to get more epidemiological and clinical information about NPEVs.
Methods
Ethical approval
This Study was approved by the Institutional Internal Review Committee. All experiments and methods were performed in accordance with approved guidelines of Internal Review Committee of National Institute of Health, Pakistan. Informed consent was obtained from the legal guardians of the children enrolled in this study.
Samples collection
Stool samples of children with Acute Flaccid Paralysis (AFP) were collected from Khyber Pakhtunkhwa (KP) and Federally Administered Tribal Areas (FATA) of Pakistan as per WHO guidelines within 14 days from the date of onset. Samples were transported to WHO Regional Reference Laboratory for Polio Eradication Initiative, Department of Virology, National Institute of Health, Islamabad, Pakistan under cold conditions along with patient clinical history and demographic data. A 60-day follow-up investigation after the onset of paralysis was carried out in those patients who experienced residual paralysis or weakness in one or more limb/s.
Virus Isolation
Stool samples were treated with chloroform in phosphate buffer saline solution (PBS) to remove bacteria and fungi. A total of 200 μl of Stool supernatant of each sample was inoculated on RD (derived from rabdomayosarcoma cells of humans) and L20B cell culture tubes (mouse lymphoma cells expressing the human poliovirus receptor) and incubated at 36 °C47,48. A daily microscopic observation of each cell culture tube was recorded for characteristic enterovirus cytopathic effect (CPE) and samples positive on RD cells but negative on L20B cells was re-passaged on L20B cell to rule out the presence of polioviruses.
Microneutralization Assay
Microneutralization assay was performed in 96-well tissue culture plates by using typing sera kit provided by National Institute for Public Health and the Environment (RIVM), Bilthoven47.
RNA Extraction, Enterovirus Detection by qRT-PCR and VP1 R T-PCR Amplification
RNA was extracted from the supernatant of infected cells using the QIAamp Viral RNA extraction Kit (Qiagen, Valencia, CA) as per the manufacturer’s instruction. Non polio enteroviruses (NPEVs) were confirmed by real time reverse transcription Polymerase Chain Reaction (qRT-PCR) targeting 5´ UTR21. All screened NPEVs were further characterized by sequencing of VP1 gene through RT-PCR. Initially, complementary DNA (cDNA) was synthesized in a 20 μL reaction mixture comprising of 11 μL of each viral RNA, 4 μL of 5x transcriptase buffer, 2 μL of 10 mM DTT, 0.5 μL of 25 mM each dNTP mix (Roche), 1 μL of 40U of RNase Inhibitor and 1 μL of 20U AMV reverse transcriptase (Thermo Scientific) at 42 °C for 45 min. For amplification of VP1 region RCR was performed by using 2 μL of cDNA, 5 μL of 10xTaq buffer, 3.5 μL of MgCl2 (50 mM), 1.0 μL of 12.5 mM dNTPs, 0.5 μL of Taq DNA polymerase (5 units/ μL), 37 μL of water and 0.5 μL of each primer (10 μM) 224–222 targeting VP3 (nt 2204–2223) and VP1 (nt 2969–2951) region of virus genome49. Thermocycler parameters for RT-PCR was 95 °C for 3 min followed by 40 cycles of 94 °C for 45 s, 42 °C for 45 s and 72 °C for 1 min. Specific RT-PCR products were purified using the Qiaquick PCR purification kit (Qiagen) and stored at −20 °C till further procedure.
Nucleotide Sequencing and Phylogenetic analysis
The purified RT-PCR products were sequenced with overlapping primer set 224–22222 using a PRISM Big Dye Terminator cycle sequencing reaction kit version 3.1 (Perkin Elmer Applied Biosystem). Sequence data was generated by an automated ABI 3130 Genetic Analyzer and sequences were assembled and edited by using Sequencher 4.9 software (GeneCodes) (http://www.genecodes.com).
Genetic differences among E-19 isolates were analyzed by MEGA 5.0 software using partial (399 bp) VP1 region50. Sequence of E-19 prototype and all the representative strains retrieved from GenBank (http://www.ncbi.nlm.nih. gov/BLAST51 and aligned using ClustalW52. Evolutionary distances have been computed by using Maximum Composite Likelihood model and Jones–Thornton–Taylor (JTT) model for nucleotide and deduced amino acid sequences respectively53. Phylogenetic tree was constructed by maximum-likelihood method54 with 1000 bootstrap replicates55,56.
The evolutionary history of Echovirus 19 dataset was estimated by using a Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in the Bayesian Evolutionary Sampling Tree (BEAST) package (v1.8.1; http://beast.bio.ed.ac.uk/Beast)57. The Hasegawa-Kishino-Yano (HKY) + Gamma distribution (G) was the best fitting model of evolution as estimated by jModelTest 2.1.758,59. Relaxed molecular clock model (the uncorrelated lognormal-distributed model [UCLD]) was employed. MCMC analyses were run for 20 million generations, sampling a tree for every 1000 steps and the first 10% were discarded as burn-in. MCMC convergence and effective sample size (ESS) estimates were checked with Tracer (v1.6.0; http://beast.bio.ed.ac.uk/Tracer). A maximum clade credibility (MCC) tree was obtained using TreeAnnotator program (v1.8.1; http://beast.bio.ed.ac.uk/TreeAnnotator) and visualized in FigTree (v1.4.2; http://beast.bio.ed.ac.uk/figtree). Time to the most recent common ancestor (tMRCA) at each node in the phylogeny was calculated from the height value in MCC tree and 95% highest posterior density (HPD) interval was estimated.
Additional Information
Accession codes: VP1 sequences of echovirus 19 strains identified in this study were deposited in GenBank under accession number KP814112 to KP814131.
How to cite this article: Angez, M. et al. Identification of new genotype of Echovirus 19 from children with Acute Flaccid Paralysis in Pakistan. Sci. Rep. 5, 17456; doi: 10.1038/srep17456 (2015).
References
Oberste, M. S., Maher, K., Kilpatrick, D. R. & Pallansch, M. A. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. Journal of virology 73, 1941–1948 (1999).
Melnick, J. L. & Hampil, B. WHO collaborative studies on enterovirus reference antisera; fourth report. Bulletin of the World Health Organization 48, 381–396 (1973).
Melnick, J. L. & Wimberly, I. L. Lyophilized combination pools of enterovirus equine antisera: new LBM pools prepared from reserves of antisera stored frozen for two decades. Bull World Health Organ 63, 543–550 (1985).
Hyypia, T., Auvinen, P. & Maaronen, M. Polymerase chain reaction for human picornaviruses. J Gen Virol 70 (Pt 12), 3261–3268 (1989).
Santti, J., Harvala, H., Kinnunen, L. & Hyypia, T. Molecular epidemiology and evolution of coxsackievirus A9. The Journal of general virology 81, 1361–1372 (2000).
Caro, V., Guillot, S., Delpeyroux, F. & Crainic, R. Molecular strategy for ‘serotyping’ of human enteroviruses. The Journal of general virology 82, 79–91 (2001).
Palacios, G., Casas, I., Tenorio, A. & Freire, C. Molecular identification of enterovirus by analyzing a partial VP1 genomic region with different methods. Journal of clinical microbiology 40, 182–192 (2002).
Adams, M. J., King, A. M. & Carstens, E. B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013). Archives of virology doi: 10.1007/s00705-013-1688-5 (2013).
Brown, B., Oberste, M. S., Maher, K. & Pallansch, M. A. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. Journal of virology 77, 8973–8984 (2003).
Knowles, N. J. et al. In King AMQ,Picornaviridae: the 9th ICTV Report and Beyond. 2012 edn, (Elsevier, 2012).
Oberste, M. S. et al. Enteroviruses 76, 89, 90 and 91 represent a novel group within the species Human enterovirus A. The Journal of general virology 86, 445–451, doi: 10.1099/vir.0.80475-0 (2005).
Oberste, S., Drescher, K. & Tracy, S. Group B coxsackieviruses. Preface. Curr Top Microbiol Immunol 323, v–vi (2008).
Rotbart, H. A. Enteroviral infections of the central nervous system. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 20, 971–981 (1995).
Sawyer, M. H. Enterovirus infections: diagnosis and treatment. Pediatr Infect Dis J 18, 1033–1039; quiz 1040 (1999).
Ramos-Alvarez, M. & Sabin, A. B. Enteropathogenic viruses and bacteria; role in summer diarrheal diseases of infancy and early childhood. J Am Med Assoc 167, 147–156 (1958).
Faulkener, R. S. a. O. & R. L. Aseptic Meningitis due to Type 19 Echo Virus Infection. New England Journal of Medicine 263, 2, doi: 10.1056/NEJM196009152631107 (1960).
Bacon, C. J. & Sims, D. G. Echovirus 19 infection in infants under six months. Arch Dis Child 51, 631–633 (1976).
Codd, A. A. & Hale, J. H. Epidemic of echovirus 19 in the north-east of England. The Journal of hygiene 76, 307–317 (1976).
Lukashev, A. N. et al. Molecular epidemiology of enteroviruses causing uveitis and multisystem hemorrhagic disease of infants. Virology 307, 45–53 (2003).
Kesson, A. M., Choo, C. M., Troedson, C., Thorley, B. R. & Roberts, J. A. Echovirus 19 associated with a case of acute flaccid paralysis. Journal of paediatrics and child health 49, E239–242, doi: 10.1111/jpc.12043 (2013).
Kilpatrick, D. R. et al. Rapid group-, serotype- and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. Journal of clinical microbiology 47, 1939–1941, doi: 10.1128/JCM.00702-09 (2009).
Nix, W. A., Oberste, M. S. & Pallansch, M. A. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. Journal of clinical microbiology 44, 2698–2704, doi: 10.1128/JCM.00542-06 (2006).
Rico-Hesse, R., Pallansch, M. A., Nottay, B. K. & Kew, O. M. Geographic distribution of wild poliovirus type 1 genotypes. Virology 160, 311–322 (1987).
Kew, O. M. & Nottay, B. K. Molecular epidemiology of polioviruses. Reviews of infectious diseases 6 Suppl 2, S499–504 (1984).
Mulders, M. N., Salminen, M., Kalkkinen, N. & Hovi, T. Molecular epidemiology of coxsackievirus B4 and disclosure of the correct VP1/2A(pro) cleavage site: evidence for high genomic diversity and long-term endemicity of distinct genotypes. The Journal of general virology 81, 803–812 (2000).
Oberste, M. S., Nix, W. A., Kilpatrick, D. R., Flemister, M. R. & Pallansch, M. A. Molecular epidemiology and type-specific detection of echovirus 11 isolates from the Americas, Europe, Africa, Australia, southern Asia and the Middle East. Virus research 91, 241–248 (2003).
Chan, Y. F., Sam, I. C. & AbuBakar, S. Phylogenetic designation of enterovirus 71 genotypes and subgenotypes using complete genome sequences. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases 10, 404–412, doi: 10.1016/j.meegid.2009.05.010 (2010).
Rao, C. D., Yergolkar, P. & Shankarappa, K. S. Antigenic diversity of enteroviruses associated with nonpolio acute flaccid paralysis, India, 2007-2009. Emerging infectious diseases 18, 1833–1840, doi: 10.3201/eid1811.111457 (2012).
Michos, A. G. et al. Aseptic meningitis in children: analysis of 506 cases. PloS one 2, e674, doi: 10.1371/journal.pone.0000674 (2007).
Dietz, V., Andrus, J., Olive, J. M., Cochi, S. & de Quadros, C. Epidemiology and clinical characteristics of acute flaccid paralysis associated with non-polio enterovirus isolation: the experience in the Americas. Bulletin of the World Health Organization 73, 597–603 (1995).
Dhole, T. N. et al. Non-polio enteroviruses in acute flaccid paralysis children of India: vital assessment before polio eradication. Journal of paediatrics and child health 45, 409–413, doi: 10.1111/j.1440-1754.2009.01529.x (2009).
Kapoor, A., Ayyagari, A. & Dhole, T. N. Non-polio enteroviruses in acute flaccid paralysis. Indian journal of pediatrics 68, 927–929 (2001).
Mehrabi, Z. et al. Molecular detection of different types of non-polio enteroviruses in acute flaccid paralysis cases and healthy children, a pilot study. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology 50, 181–182, doi: 10.1016/j.jcv.2010.10.006 (2011).
Saeed, M. et al. Epidemiology and clinical findings associated with enteroviral acute flaccid paralysis in Pakistan. BMC infectious diseases 7, 6, doi: 10.1186/1471-2334-7-6 (2007).
Shaukat, S. et al. Identification and characterization of unrecognized viruses in stool samples of non-polio acute flaccid paralysis children by simplified VIDISCA. Virol J 11, 146, doi: 10.1186/1743-422X-11-146 (2014).
Shaukat, S. et al. Characterization of non-polio enterovirus isolates from acute flaccid paralysis children in Pakistan reflects a new genotype of EV-107. Virus research 170, 164–168, doi: 10.1016/j.virusres.2012.09.010 (2012).
Angez, M. et al. Molecular epidemiology of enterovirus B77 isolated from non polio acute flaccid paralytic patients in Pakistan during 2013. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases 29, 189–195, doi: 10.1016/j.meegid.2014.11.021 (2015).
Figueroa, J. P., Ashley, D., King, D. & Hull, B. An outbreak of acute flaccid paralysis in Jamaica associated with echovirus type 22. Journal of medical virology 29, 315–319 (1989).
Zhu, Z. et al. Molecular epidemiological analysis of echovirus 19 isolated from an outbreak associated with hand, foot and mouth disease (HFMD) in Shandong Province of China. Biomedical and environmental sciences : BES 20, 321–328 (2007).
Saraswathy, T. S. et al. Laboratory acute flaccid paralysis surveillance in Malaysia: a decade of commitment to the WHO global polio eradication initiative. The Southeast Asian journal of tropical medicine and public health 35, 421–424 (2004).
Oyero, O. G. & Adu, F. D. Non-polio enteroviruses serotypes circulating in Nigeria. Afr J Med Med Sci 39 Suppl, 201–208 (2010).
Tang, J. et al. Molecular epidemiology and recombination of human enteroviruses from AFP surveillance in Yunnan, China from 2006 to 2010. Sci Rep 4, 6058, doi: 10.1038/srep06058 (2014).
Starlin, R. et al. Acute flaccid paralysis syndrome associated with echovirus 19, managed with pleconaril and intravenous immunoglobulin. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 33, 730–732, doi: 10.1086/322624 (2001).
Palacios, G. et al. Molecular epidemiology of echovirus 30: temporal circulation and prevalence of single lineages. Journal of virology 76, 4940–4949 (2002).
Kunkel, U. & Schreier, E. Characterization of echovirus 25 (ECV 25) in the VP1/2A gene junction region. Brief report. Archives of virology 144, 2253–2258 (1999).
GPEI. (The Polio Eradication Initiative, 2013).
WHO. (World Health Organization (WHO/IVB/04.10), Geneva 27 Switzerland, 2004).
Melnick, J. L. et al. in Virology Vol. 3rd (ed Knipe, D. M., Fields, B. N., Howley, P. M., Chanock, R. M., Melnick, J. L., Monath, T. P., Roizman, B. & Straus, S. E. ) 655–712 (Lippincott-Raven, 1996).
Oberste, M. S. et al. Comparison of classic and molecular approaches for the identification of untypeable enteroviruses. Journal of clinical microbiology 38, 1170–1174 (2000).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular biology and evolution 24, 1596–1599, doi: 10.1093/molbev/msm092 (2007).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. Journal of molecular biology 215, 403–410, doi: 10.1016/S0022-2836(05)80360-2 (1990).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680 (1994).
Jones, D. T., Taylor, W. R. & Thornton, J. M. The rapid generation of mutation data matrices from protein sequences. Computer applications in the biosciences : CABIOS 8, 275–282 (1992).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Molecular biology and evolution 28, 2731–2739, doi: 10.1093/molbev/msr121 (2011).
Pattengale, N. D., Alipour, M., Bininda-Emonds, O. R., Moret, B. M. & Stamatakis, A. How many bootstrap replicates are necessary? J Comput Biol 17, 337–354, doi: 10.1089/cmb.2009.0179 (2010).
Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 39, 783–791 (1985).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular biology and evolution 29, 1969–1973, doi: 10.1093/molbev/mss075 (2012).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nature methods 9, 772, doi: 10.1038/nmeth.2109 (2012).
Guindon, S. & Gascuel, O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology 52, 696–704 (2003).
Acknowledgements
The authors gratefully acknowledge World Health Organization AFP surveillance staff working in Pakistan for their technical support.
Author information
Authors and Affiliations
Contributions
S.Z. Conceived, designed and supervised the study. M.A. Conceived the study, performed experiments and drafted the paper. S.S. Performed evolutionary analysis and contributed to the writing. Sal.S. Helped to interpret results. M.M.A. Analyzed Data. A.K. Contributing in performing experiments. S.R. Gathered and analyzed the data. R.Z. Designed the study and contributed to the writing. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Angez, M., Shaukat, S., Zahra, R. et al. Identification of new genotype of Echovirus 19 from children with Acute Flaccid Paralysis in Pakistan. Sci Rep 5, 17456 (2015). https://doi.org/10.1038/srep17456
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep17456
This article is cited by
-
Genetic Characteristics and Phylogeographic Dynamics of Echovirus
Journal of Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.