Abstract
Fusicoccane diterpenoids usually possess a fused 5-8-5 tricyclic ring system, which are biogenetically generated from geranylgeranyl diphosphate (GGDP). In our report, three novel diterpenoid alkaloids with fusicoccane skeleton, pericolactines A–C (1–3), were isolated from Periconia sp.. Their structures with absolute configurations were determined by spectroscopic analyses and quantum chemical ECD calculation. Pericolactines A–C (1–3) are a new class of diterpenoid alkaloids with an unusual fused 5-5-8-5 tetracyclic ring system, which derive from a geranylgeranyl diphosphate (GGDP) and serine conjugated biosynthesis. They belong to the atypical diterpenoid alkaloids.
Similar content being viewed by others
Introduction
Diterpenoid alkaloids are a kind of nitrogen-containing diterpenes that derive from a terpene-amino acid conjugated biosynthesis. According their structural characteristics, the majority of them are classified as typical diterpenoid alkaloids, including C18-, C19- and C20-diterpenoid alkaloids1,2,3,4. Till now, only a few atypical diterpenoid alkaloids have been reported, such as concavine, chamobtusin A and haterumaimides4.
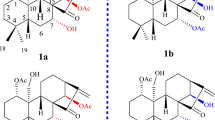
During our ongoing research on bioactive secondary metabolites from fungi5,6,7,8,9,10,11,12,13, a chemical investigation on metabolites from Periconia sp. (No. 19-4-2-1) isolated from the lichen Parmelia sp. was carried out, which led to the isolation of three novel polycyclic diterpenoid alkaloids, pericolactines A–C (1–3). Pericolactines A–C (1–3) are a new class of diterpenoid alkaloids featuring a fused 5-5-8-5 tetracyclic skeleton and belong to atypical diterpenoid alkaloids, which derive from the terpene-amino acid conjugated biosynthesis. Details of the structure elucidation for 1–3 (Fig. 1) are reported herein.
Results
Pericolactine A (1), isolated as a white amorphous powder, was assigned the molecular formula C24H35NO5 (eight degrees of unsaturation) according to a quasi-molecular ion at m/z 418.2595 [M + H]+ in its HRESIMS spectrum. The 13C NMR spectrum (Table 1) showed 24 carbon signals, which was consistent with the deduction of the HRESIMS. Combined with the DEPT-135 experiment, these carbons can be categorized into six sp2 quaternary carbons [including two carbonyl carbons (δC 178.1 and 172.7) and four olefinic carbons], one sp3 quaternary carbon (δC 52.0), five sp3 methine carbons, eight sp3 methylene carbons and four methyl carbons (δC 28.5, 20.8, 15.8 and 12.1, respectively). In the 1H NMR spectrum of 1 (Table 1), the characteristic protons for four methyl groups [δH 1.87 (s), 1.11 (s), 1.03 (d, J = 7.1 Hz) and 0.98 (d, J = 7.0 Hz)] were observed. All the proton resonances were assigned to relevant carbon atoms through the HSQC experiment. The analysis of the 1H-1H COSY experiment revealed the presence of five isolated spin systems (C-1–C-2–C-3(C-16)–C-4–C-5, C-8–C-9, C-12–C-13, C-19–C-15–C-20 and C-1′–C-2′) as shown in Fig. 2. Combined with the 1H-1H COSY analysis and the degrees of unsaturation, the HMBC correlations from Ha-1/Hb-1 to C-6, from H-2 to C-7, from H-4 to C-6, from H-5 to C-6/C-7/C-17, from Ha-8/Hb-8 to C-6/C-7/C-10/C-17, from Ha-9/Hb-9 to C-7/C-10/C-11, from Ha-12/Hb-12 to C-14, from Ha-13/Hb-13 to C-10, from H3-16 to C-2/C-3/C-4, from H3-18 to C-1/C-10/C-11/C-12 revealed a 5-8-5 fused ring system (rings A/B/C, Fig. 2). With the 1H-1H COSY correlations between Ha-19/Hb-19/H3-20 and H-15, the HMBC correlations from H-15 to C-10/C-13/C-14, from Ha-19/Hb-19 to C-14/C-15/C-20/19-OCOCH3, H3-20 to C-14/C-15/C-19 and from 19-OCOCH3 to 19-OCOCH3 revealed a 1-acetoxypropan-2-yl located at C-14. With the 1H-1H COSY correlations between Ha-1′/Hb-1′ and Ha-2′/Hb-2′, the key HMBC correlations from Ha-1′/Hb-1′ to C-5/C-17 revealed a γ-lactam ring in 1 (ring D, Fig. 2). On the basis of the analyses of 1H-1H COSY, HMBC, the degrees of unsaturation and the molecular formula, the planar structure of 1 was deduced as shown in Fig. 2 and the assignments of all proton and carbon resonances are shown in Table 1.
The ROESY correlations between H-2 and H-4/H-5 and between H-3 and H-5 signified that H-2, H-3, H-4 and H-5 located on the same face of the ring A (Fig. 3). Furthermore, the ROESY correlation between H-2 and Ha-12 signified that H-2 and C-12 located on the same face of the ring B (Fig. 3), while H3-18 was on the other face. Combined with the ROESY correlations between H-5 and 19-OCOCH3, between Ha-9 and H-15, between Hb-13 and Ha-19/Hb-19 and between Ha-13/Hb-13 and H3-20, the relative configurations of C-2, C-3, C-4, C-5, C-11 and C-15 in 1 were assigned as 2S*, 3R*, 4R*, 5S*, 11R* and 15R*, respectively (Fig. 3). Thus, the structure of 1 was established as a new diterpenoid alkaloids with fusicoccane skeleton.
Pericolactine B (2) was obtained as a white amorphous powder. It was assigned the molecular formula C22H33NO4 (seven degrees of unsaturation) according to a quasi-molecular ion at m/z 376.2491 [M + H]+ in its HRESIMS spectrum. The molecular weight of 2 was a 42 atomic mass unit (C2H2O) less than 1, which indicated that 2 may be a 19-deacetylated derivative of 1. The 1H and 13C NMR spectra of 2 showed resonances very similar to 1, except for the disappearance of one acetyl group. Further detailed NMR analyses involving 1H-1H COSY and HMBC spectra (see Supplementary Table S2 online) confirmed the above deduction and given the assignments of all proton and carbon resonances (Table 1).
The fact that 2 is the deacetylated derivative of 1 was confirmed by acid hydrolysis. Pericolactine A (1) was treated with H2SO4 in MeOH and then the product prepared from 1 was compared with 2 using HPLC (see Supplementary Figure S1 online), which displayed that the retention times of the product prepared from 1 were identical to 2 isolated from fungal broth in three eluting systems. Based on the above mentioned fact, the relative configuration of 2 was assigned as 2S*, 3R*, 4R*, 5S*, 11R*, 15R*, which was the same as 1. The absolute configurations of C-2, C-3, C-4, C-5, C-11 and C-15 in 2 were determined by quantum chemical ECD calculation. The conformational analysis for a pair of enantiomers ((2S, 3R, 4R, 5S, 11R, 15R)-2 and (2R, 3S, 4S, 5R, 11S, 15S)-2) was carried out in CONFLEX version 7.0 with an energy window for acceptable conformers (0–3 kcal mol−1). The acceptable conformers were obtained and continued to be optimized in Gaussian09. After that, five lowest energy conformers were found out. These lowest energy conformers (Fig. 4) were submitted to the ECD calculation at [B3P86/6-311++G (2d, p)] level and the predicted ECD curve of (2S, 3R, 4R, 5S, 11R, 15R)-2 was similar to the experimental one (Fig. 5 and see Supplementary information). Therefore, the absolute configuration of 2 was established as 2S, 3R, 4R, 5S, 11R and 15R.
Since 1 and 2 possess the similar ECD curves (Fig. 5) and 1 and 2 coexist in the same strain, 1 and 2 possess the same absolute configurations. Thus, the absolute configuration of 1 was also assigned as 2S, 3R, 4R, 5S, 11R and 15R.
Pericolactine C (3) was isolated as a white amorphous powder. Its molecular formula was established as C23H35NO5 (seven degrees of unsaturation) by the quasi-molecular ion at m/z 428.2418 [M + Na]+ in the HRESIMS. The 1H and 13C NMR spectra of 3 were very similar to 2, expect for the absence of C-5 methine and the appearance of an oxygenated sp3 quaternary carbon (δC 97.0) and a methoxy group (δC 50.6/δH 2.99). The key HMBC correlation from the additional methoxy group at δH 2.99 to C-5 (δC 97.0) indicated that H-5 in 2 was substituted by methoxy group in 3. On the basis of 2D NMR analysis (see Supplementary Table S3 online), the planar structure of 3 was established (Fig. 2) and the assignments of all proton and carbon resonances are shown in Table 1.
The relative configuration of 3 was elucidated by analysis of the ROESY experiment. The ROESY correlations between 5-OCH3 and H-2/H-3 signified that H-2, H-3 and 5-OCH3 located on the same face of the ring A (Fig. 6). Furthermore, the ROESY correlation between H-2 and Ha-12 signified that H-2 and C-12 located on the same face of the ring B (Fig. 6), while the H3-18 was on the other face. Combined with the ROESY correlations between 5-OCH3 and Ha-19/Hb-19 and between Ha-19/Hb-19/H3-20 and Ha-13/Hb-13, the relative configurations of C-2, C-3, C-5, C-11 and C-15 were assigned as 2S*, 3R*, 5R*, 11R* and 15R* (Fig. 6). However, the coupling constant between H-3 and H-4 (3JH-3, H-4 = 8.8 Hz) of 3 was different from 1 and 2 (3JH-3, H-4 = 3.7 Hz), which suggested that the epimerization was at C-4 in 3. Therefore, the relative configuration of 3 was determined as 2S*, 3R*, 4S*, 5R*, 11R* and 15R*. The absolute configuration of 3 was determined by quantum chemical ECD calculation. The conformational analysis for a pair of enantiomers ((2S,3R,4S,5R,11R,15R)-3 and (2R,3S,4R,5S,11S,15S)-3) was carried out in CONFLEX version 7.0 with an energy window for acceptable conformers (0–3 kcal mol−1). The acceptable conformers were obtained and continued to be optimized in Gaussian09. After that, only one lowest energy conformer was found out. The lowest energy conformer was submitted to the ECD calculation at [B3P86/6-311++G (2d, p)] level and the predicted ECD curve of (2S, 3R, 4S, 5R, 11R, 15R)-3 was similar to the experimental one (Fig. 7 and see Supplementary information). On the basis of the above analyses, the absolute configuration of 3 was assigned as 2S, 3R, 4S, 5R, 11R and 15R.
All isolated compounds were subjected to a paper disk-diffusion assay14,15 for antimicrobial activities against two bacteria (Staphylococcus aureus 209P and Escherichia coli ATCC0111) and two fungi (Candida albicans FIM709 and Aspergillus niger R330). In addition, all isolated compounds were also evaluated by MTT method16,17 for their cytotoxicity against five human tumor cell lines, including HL-60, SMMC-7721, A-549, MCF-7 and SW480, with cisplatin and paclitaxel as the positive controls. However, compounds showed no potent activity (see Supplementary Table S4 and S5 online).
Discussion
Fusicoccane diterpenoids usually possessing a tricyclic (5-8-5) ring system (such as brassicicenes, cyclooctatins, fusicoccins and periconicins) are biogenetically generated from geranylgeranyl diphosphate (GGDP)18, which are found from various natural sources, including bacteria19,20, fungi21,22,23,24, liverworts25,26, algas27 and higher plants28,29. Fusicoccane diterpenoids exhibit diverse biological activities, such as plant growth regulating activity (fusicoccins)30, lysophospholipase inhibitory activity (cyclooctatin)31, antimicrobial activity (periconicins)32,33, nitrification inhibitory activity (brachialactone)34, cytotoxicity against tumor cells (cotylenins)35, inhibiting insulin-stimulated GLUT4 fusion activity (fusicoccins)35 and so on.
Pericolactines A–C (1–3) are the first nitrogen-containing fusicoccane diterpenoids, which derive from fusicocca-2,10(14)-diene and serine (Figure 8). Due to the participation of serine in the biogenetic pathway, pericolactines A–C (1–3) represent a new class of diterpenoid alkaloids.
Materials and Methods
General experimental procedures
Optical rotations were measured on a JASCO P1020 digital polarimeter and UV data were obtained with a JASCO V-550 UV/vis spectrometer. The CD spectra were recorded in MeOH using a JASCO J-810 spectrophotometer at room temperature. IR data were recorded using JASCO FT/IR-480 Plus spectrometer. HRESIMS spectra were obtained on Waters Synapt G2 TOF mass spectrometer. The NMR data were acquired with a Bruker AV 400 NMR spectrometer using solvent signals (CD3OD: δH 3.30/δC 49.0) as standards. Column chromatography (CC) was carried out on Sephadex LH-20 (Pharmacia, USA) and ODS (60–80 μm, YMC). TLC was performed on precoated silica gel plate (SGF254, 0.2 mm, Yantai Chemical Industry Research Institute, China). Analytical HPLC was performed on a Dionex HPLC system equipped with an Ultimate 3000 pump, an Ultimate 3000 diode array detector, an Ultimate 3000 column compartment, an Ultimate 3000 autosampler (Dionex, USA) and an Alltech (Grace) 2000ES evaporative light scattering detector (Alltech USA) using a Phenomenex Gemini C18 column (4.6 × 250 mm, 5 μm). Preparative HPLC was carried out on Shimadzu LC-6AD system equipped with UV detectors, using a Phenomenex Gemini C18 column (21.2 × 250 mm, 5 μm). Semi-preparative HPLC was carried out on Shimadzu LC-6AD system equipped with UV detectors, using a YMC-Pack ODS-A column (10.0 × 250 mm, 5 μm).
Fungus material
The strain of Periconia sp. (No. 19-4-2-1) was isolated by one of the authors (L.D. Guo) from the lichen Parmelia sp. collected from Changbai Mountain, Jilin Province, China, in August 2006. The fungus strain was identified as Periconia sp. based on the morphological characteristics and sequence analysis of the internal transcribed spacer (ITS) regions ITS1-5.8S-ITS2 (GenBank accession No. KP873157). Briefly, the genomic DNA of the fungus was extracted and used as a template for amplification of ITS region by fungus-specific universal primer pair ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′). The resulting DNA fragment was sequenced and deposited at GenBank. Species were identified by searching databases using the BLAST sequence analysis tool (http://www.ncbi.nlm.nih.gov/BLAST/). The strain was assigned the accession number 19-4-2-1 in the culture collection at the Institute of Traditional Chinese Medicine and Natural Products, college of Pharmacy, Jinan University, Guangzhou. The fungus was cultured on slants of potato dextrose agar at 25 °C for 5 days. Agar plugs were used to inoculate four Erlenmeyer flasks (250 mL), each containing 100 mL of potato dextrose broth. Four flasks of the inoculated media were incubated at 25 °C on a rotary shaker at 200 rpm for 5 days to prepare the seed culture. Fermentation was carried out in 20 Erlenmeyer flasks (500 mL), each containing 70 g of rice. Distilled H2O (105 mL) was added to each flask and the rice was soaked overnight before autoclaving at 120 °C for 30 min. After cooling to room temperature, each flask was inoculated with 5.0 mL of the spore inoculum and incubated at room temperature for 45 days.
Extraction and isolation
The culture was extracted thrice with EtOAc and the organic solvent was evaporated to dryness under vacuum to afford a crude extract (33.4 g). The crude extract was dissolved in 90% v/v aqueous MeOH (500 mL) and partitioned against the same volume of cyclohexane to afford a cyclohexane fraction (C, 24.5 g) and an aqueous MeOH fraction (W, 8.7 g). The aqueous MeOH fraction (W, 8.7 g) was separated by ODS CC eluting with MeOH-H2O (30:70, 50:50, 70:30 and 100:0, v/v) to afford four fractions (W1 to W4). Fraction W3 (2.6 g) was further separated on a ODS column with a gradient of MeOH-H2O (55:45, 60:40, 65:35, 70:30 and 100:0, v/v) to give seven subfractions (W3a to W3 g). Subfraction W3c (1.3 g) was subjected to Sephadex LH-20 CC using MeOH to afford four portions (W3c1 to W3c4). W3c3 (994.5 mg) was separated on preparative HPLC using CH3CN-H2O (35:65, v/v) to yield 1 (6.8 mg) and 3 (4.4 mg). Fraction W2 (2.3 g) was also subjected to a ODS column with a gradient of MeOH-H2O (35:65, 40:60, 45:55, 50:50, 55:45 and 100:0, v/v) to give eight subfractions (W2a to W2h). Subfraction W2f (399.6 mg) was separated by ODS CC eluting with MeOH-H2O (50:50, v/v) to afford three portions (W2f1 to W2f3). W2f2 (299.0 mg) was purified on semi-preparative HPLC by using CH3CN-H2O (35: 65, v/v) to yield 2 (11.2 mg).
Spectroscopic data of 1–3
Pericolactine A (1): white amorphous powder;  −29.1 (c 1.0, MeOH); UV (MeOH) λmax (log ε) 207 (3.97) nm; CD (c 2.7 × 10−4 M, MeOH) λmax (Δε) 220 (−5.17), 245 (+3.10); IR (KBr) νmax 3437, 2944, 1718, 1658, 1385, 1240, 1038 cm−1; HRESI-TOF-MS m/z 418.2595 [M + H]+ (calcd for C24H36NO5, 418.2593); The 1H and 13C NMR data, see Supplementary Table S1 online.
−29.1 (c 1.0, MeOH); UV (MeOH) λmax (log ε) 207 (3.97) nm; CD (c 2.7 × 10−4 M, MeOH) λmax (Δε) 220 (−5.17), 245 (+3.10); IR (KBr) νmax 3437, 2944, 1718, 1658, 1385, 1240, 1038 cm−1; HRESI-TOF-MS m/z 418.2595 [M + H]+ (calcd for C24H36NO5, 418.2593); The 1H and 13C NMR data, see Supplementary Table S1 online.
Pericolactine B (2): white amorphous powder;  −33.5 (c 1.0, MeOH); UV (MeOH) λmax (log ε) 209 (3.86) nm; CD (c 3.4 × 10−4 M, MeOH) λmax (Δε) 220 (−6.48), 245 (+4.10); IR (KBr) νmax 3390, 2941, 1656, 1034 cm−1; HRESI-TOF-MS m/z 376.2491 [M + H]+ (calcd for C22H34NO4, 376.2488); The 1H and 13C NMR data, see Supplementary Table S2 online.
−33.5 (c 1.0, MeOH); UV (MeOH) λmax (log ε) 209 (3.86) nm; CD (c 3.4 × 10−4 M, MeOH) λmax (Δε) 220 (−6.48), 245 (+4.10); IR (KBr) νmax 3390, 2941, 1656, 1034 cm−1; HRESI-TOF-MS m/z 376.2491 [M + H]+ (calcd for C22H34NO4, 376.2488); The 1H and 13C NMR data, see Supplementary Table S2 online.
Pericolactine C (3): white amorphous powder;  −27.6 (c 1.0, MeOH); UV (MeOH) λmax (log ε) 207 (4.04) nm; CD (c 3.0 × 10−4 M, MeOH) λmax (Δε) 226 (−8.09), 259 (+1.29); IR (KBr) νmax 3403, 2941, 1681, 1385, 1050 cm−1; HRESI-TOF-MS m/z 428.2418 [M + Na]+ (calcd for C23H35NO5Na, 428.2413); The 1H and 13C NMR data, see Supplementary Table S3 online.
−27.6 (c 1.0, MeOH); UV (MeOH) λmax (log ε) 207 (4.04) nm; CD (c 3.0 × 10−4 M, MeOH) λmax (Δε) 226 (−8.09), 259 (+1.29); IR (KBr) νmax 3403, 2941, 1681, 1385, 1050 cm−1; HRESI-TOF-MS m/z 428.2418 [M + Na]+ (calcd for C23H35NO5Na, 428.2413); The 1H and 13C NMR data, see Supplementary Table S3 online.
Acid hydrolysis of 1
Compound 1 (1.0 mg) stirred with 98% H2SO4 (2 μL) in MeOH (2 mL) at 40 °C for 3.5 h. After neutralization with ammonia, the solvent was evaporated to yield the mixture. Then the mixture was compared with 2 by HPLC, which displayed that the retention time of the product prepared from compound 1 was identical to 2 isolated from fungal broth (see Supplementary Figure S1 online).
Additional Information
How to cite this article: Wu, Y.-H. et al. Pericolactines A–C, a New Class of Diterpenoid Alkaloids with Unusual Tetracyclic Skeleton. Sci. Rep. 5, 17082; doi: 10.1038/srep17082 (2015).
References
Pelletier, S. W. & Page, S. W. Diterpenoid alkaloids. Nat Prod Rep 1, 375–386 (1984).
Pelletier, S. W. & Page, S. W. Diterpenoid alkaloids. Nat Prod Rep 3, 451–464 (1986).
Atta-ur-Rahman & Choudhary, M. I. Diterpenoid and steroidal alkaloids. Nat Prod Rep 16, 619–635 (1999).
Wang, F. P., Chen, Q. H. & Lu, X. Y. Diterpenoid alkaloids. Nat Prod Rep 27, 529–570 (2010).
Chen, G. D. et al. Xanthoquinodins from the endolichenic fungal strain Chaetomium elatum. J Nat Prod 76, 702–709 (2013).
Ye, F. et al. Xinshengin, the first altenusin with tetracyclic skeleton core from Phialophora spp. Tetrahedron Lett 54, 4551–4554 (2013).
Zheng, Q. C. et al. Nodulisporisteriods A and B, the first 3,4-seco-4-methyl-progesteroids from Nodulisporium sp. Steroids 78, 896–901 (2013).
Wu, Z. Y. et al. Xylariterpenoids A-D, four new sesquiterpenoids from the Xylariaceae fungus. RSC Adv 4, 54144–54148 (2014).
Xiong, H. et al. Sporormiellin A, the first tetrahydrofuran-fused furochromone with an unprecedented tetracyclic skeleton from Sporormiella minima. RSC Adv 4, 24295–24299 (2014).
Zhao, Q. et al. Nodulisporiviridins A-H, bioactive viridins from Nodulisporium sp.. J Nat Prod 78, 1221–1230 (2015).
Zhao, Q. et al. Nodulisporisteroids C-L, new 4-methyl-progesteroid derivatives from Nodulisporium sp. steroids 102, 101–109 (2015).
Bao, Y. R. et al. 4,5-seco-Probotryenols A-C, a new type of sesquiterpenoids from Stachybotrys bisbyi. RSC Adv 5, 46252–46259 (2015).
Wu, Y. H. et al. Pericoterpenoid A, a new bioactive cadinane-type sesquiterpene from Periconia sp.. J Asian Nat Prod Res 17, 671–675 (2015).
Groblacher, B., Maier, V., Kunert, O. & Bucar, F. J Nat Prod 75, 1393–1399 (2012).
Shen, C. C., Syu, W. J., Li, S. Y., Lin, C. H., Lee, G. H. & Sun, C. M. J Nat Prod 65, 1857–1862 (2002).
Mosmman, T. J Immunol Methods 65, 55–63 (1983).
Alley, M. C., Scudiero, D. A., Monks, A., Hursey, M. L., Czerwinski, M. J. & Fine, D. L. Cancer Res 48, 589–601 (1988).
Arens, J., Engels, B., Klopries, S., Jennewein, S., Ottmann, C. & Schulz, F. Exploration of biosynthetic access to the shared precursor of the fusicoccane diterpenoid family. Chem Commun 49, 4337–4339 (2013).
Aoyama, T., Naganawa, H., Muraoka, Y., Aoyagi, T. & Takeuchi, T. The structure of cyclooctatin, a new inhibitor of lysophospholipase. J Antibiot 45, 1703–1704 (1992).
Kawamura, A., Iacovidou, M., Hirokawa, E., Soll, C. E. & Trujillo, M. 17-Hydroxycyclooctatin, a fused 5-8-5 ring diterpene, from Streptomyces sp. MTE4a. J Nat Prod 74, 492–495 (2011).
Billio, A., Chain, E. B., de Leo, P., Erlanger, B. F., Mauri, M. & Tonolo, A. Fusicoccin: a new wilting toxin produced by Fusicoccum amygdali Del. Nature 203, 297 (1964).
Sassa, T. Cotylenins, leaf growth substances produced by a fungus Part I. isolation and characterization of cotylenins A and B. Agric Biol Chem 35, 1415–1418 (1971).
MacKinnon, S. L., Keifer, P. & Ayer, W. A. Components from the phytotoxic extract of Alternaria brassicicola, a black spot pathogen of canola. Phytochemistry 51, 215–221 (1999).
Takekawa, H., Tanaka, K., Fukushi, E., Matsuo, K. & Nehira, T. Roussoellols A and B, tetracyclic fusicoccanes from Roussoella hysterioides. J Nat Prod 76, 1047–1051 (2013).
Hashimoto, T., Tori, M., Taira, Z. & Asakawa, Y. New highly oxidized fusicoccane diterpenoids from the liverwort Plagiochila acathophylla subsp. Japonica. Tetrahedron Lett 26, 6473–6476 (1985).
Liu, N., Guo, D. X., Wang, S. Q., Wang, Y. Y., Zhang, L., Li, G. & Lou, H. X. Bioactive sesquiterpenoids and diterpenoids from the liverwort Bazzania albifolia. Chem Biodivers 9, 2254–2261 (2012).
Enoki, N., Furusaki, A., Suehiro, K., Ishida, R. & Matsumoto, T. Epoxydictymene, a new diterpene from the brown alga Dictyota dichotoma. Tetrahedron Lett 24, 4341–4342 (1983).
Okogun, J. I., Adesomoju, A. A., Adesida, G. A., Lindner H. J. & Habermehl, G. Roseanolone: a new diterpene from Hypoestes rosea. Z Naturforsch Sec C 37c, 558–561 (1982)
Adesomoju, A. A., Okogun, J. I., Cava, M. P. & Carroll, P. J. Roseadione, a diterpene ketone from Hypoestes rosea. Phytochemsitry 22, 2535–2536 (1983).
Marre, E. Fusicoccin: a tool in plant physiology. Annu Rev Plant Physiol 30, 273–288 (1979).
Kim, S. Y. et al. Cloning and heterologous expression of the cyclooctatin biosynthetic gene cluster afford a diterpene cyclase and two P450 hydroxylases. Chem Biol 16, 736–743 (2009).
Kim, S., Shin, D. S., Lee, T. & Oh, K. B. Periconicins, two new fusicoccane diterpenes produced by an endophytic fungus Periconia sp. with antibacterial activity. J Nat Prod 67, 448–450 (2004).
Shin, D. S., Oh, M. N., Yang, H. C. & Oh, K. B. Biological characterization of periconicins, bioactive secondary metabolites, produced by Periconia sp. OBW-15. J Microbiol Biotechnol 15, 216–220 (2005).
Subbarao, G. V. et al. Evidence for biological nitrification inhibition in Brachiaria pastures. Proc Natl Acad Sci U.S.A. 106, 17302–17307 (2009).
de Boer, A. H. & Leeuwen, I. J. D. Fusicoccanes: diterpenes with surprising biological functions. Trends Plant Sci 17, 360–368 (2012).
Acknowledgements
This work was financially supported by grants from the Ministry of Science and Technology of China (2012ZX09301002-003001006), the National Natural Science Foundation of China (81422054, 81373306), the Guangdong Natural Science Funds for Distinguished Young Scholar (S2013050014287), Guangdong Special Support Program (2014TQ01R420) and Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (Hao Gao, 2014). The ECD calculations were supported by the high-performance computing platform of Jinan University.
Author information
Authors and Affiliations
Contributions
H.G. and X.-S.Y. initiated the project. H.G. designed and coordinated the project. Y.-H.W. and G.-D.C. performed the extraction, isolation and structural identification of the compounds. R.-R.H. and G.-Q.W. performed the paper disk-diffusion assay and cytotoxicity assay. R.-R.H. also analyzed the data of the biological assays. G.-D.C. performed the quantum chemical ECD calculation. C.-X.W. performed the fermentation of the fungal strain (No. 19-4-2-1). D.H. conducted the sequence analysis of the fungal strain (No. 19-4-2-1). L.-D.G. supplied the strain of Periconia sp. (No. 19-4-2-1) and the morphometric identification. All authors approved the final version of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wu, YH., Chen, GD., He, RR. et al. Pericolactines A–C, a New Class of Diterpenoid Alkaloids with Unusual Tetracyclic Skeleton. Sci Rep 5, 17082 (2015). https://doi.org/10.1038/srep17082
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep17082
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.