Abstract
The reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) by defined transcription factors has been a well-established technique and will provide an invaluable resource for regenerative medicine. However, the low reprogramming efficiency of human iPSC is still a limitation for clinical application. Here we showed that the reprogramming potential of human dental pulp cells (DPCs) obtained from immature teeth is much higher than those of mature teeth DPCs. Furthermore, immature teeth DPCs can be reprogrammed by OCT3/4 and SOX2, conversely these two factors are insufficient to convert mature teeth DPCs to pluripotent states. Using a gene expression profiles between these two DPC groups, we identified a new transcript factor, distal-less homeobox 4 (DLX4), which was highly expressed in immature teeth DPCs and significantly promoted human iPSC generation in combination with OCT3/4, SOX2 and KLF4. We further show that activation of TGF-β signaling suppresses the expression of DLX4 in DPCs and impairs the iPSC generation of DPCs. Our findings indicate that DLX4 can functionally replace c-MYC and supports efficient reprogramming of immature teeth DPCs.
Similar content being viewed by others
Introduction
Previous reports showed that induced pluripotent stem cells (iPSCs) can be generated from various types of somatic cells using a combination of transcription factors, such as OCT3/4, SOX2, KLF4, c-MYC, NANOG and LIN281,2,3,4,5,6,7,8,9,10,11,12,13,14. Although these reports indicated that iPSCs can be induced from many types of somatic cells, the backgrounds of the donor cells, such as gene expression patterns and the epigenetic state, have a significant impact on the reprogramming efficiency, kinetics and the transcription factors required for the induction3,5,7,13,14,15,16. Because reprogramming of differentiated cells involves gradual changes in gene expression patterns, cell morphology and epigenetic state towards the embryonic state17,18,19,20,21, it has been assumed that stem cells and progenitor cells are more amenable to reprogramming than that of more differentiated cells5,12,16,22,23. For example, mouse and human neural stem cells can be reprogrammed with the single transcription factor OCT3/416,24. Similarly, Eminli et al. reported that hematopoietic stem and progenitor cells gave rise to iPSCs up to 300 times more efficiently than terminally differentiated B and T cells did, suggesting that the differentiation state of the starting cells affects reprogramming efficiency23. However, these stem/progenitor cells are generally inaccessible, require complicated procedures to obtain and are difficult to propagate in vitro.
Human dental pulp cells (DPCs) are obtained easily from adult teeth discarded as medical waste and contain abundant stem like cells25,26,27,28,29. In addition, DPCs can be expanded in a simple culture condition and show continuous growth for up to 30 passages28. In a previous study, we established more than 150 DPC lines isolated from extracted wisdom teeth and evaluated the potential of DPCs for iPS cell banking14,28. The donor teeth of these DPC lines were categorized into three developmental stages: crown completed (CC), root forming (RF) and root completed (RC). We randomly selected six lines isolated from various developmental stages to induce iPSCs and found that five DPC lines showed higher reprogramming efficiency than human dermal fibroblasts (HDFs). In this study, it remained unclear why DP75, isolated from the oldest patient in the RC stage, showed remarkably low reprogramming efficiency similar to HDFs.
Here, we focused on the relationship between the developmental stages of DPCs and reprogramming efficiency. We found that DPCs in the mature (RC) stage were reprogrammed less efficiently than DPCs in the immature (CC and RF) stages.
Results
DPCs isolated from immature teeth can be reprogrammed into iPSCs using two factors
First, we attempted to generate iPSCs from DP31 cells, a cell line isolated from the immature (CC) stage of teeth, using as few transcription factors as possible. We found that only two factors, OCT3/4 and SOX2 (OS), are required to induce ES-cell-like colonies repeatedly, albeit with very low frequency compared to the combination of OCT3/4, SOX2 and KLF4 (OSK) (Fig. 1a). Similarly, we obtained iPSCs from other immature tooth DPCs, such as DP1 (CC stage) and DP87 (RF stage), with only two factors (data not shown). Genomic PCR was employed to demonstrate that OS-induced iPSCs derived from DP31 cells (iPS-DP31-OS) contained only OCT3/4 and SOX2 transgenes (Fig. 1b). We also confirmed that iPS-DP31-OS cells could be expanded easily and expressed human specific pluripotency markers (Fig. 1c). These cells differentiated into all the three germ layers in vitro and in vivo (Fig. 1d). However, we did not obtain any ES-cell-like colonies from DP75, a line obtained from the mature (RC) stage of the tooth, using the OS cocktail (Fig. 1a). These results compelled us to further examine the maturation stage dependency of DPC reprogramming.
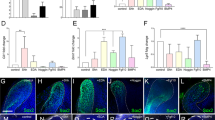
DP31 cells can be reprogrammed to iPSCs using OCT3/4 and SOX2.
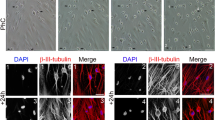
(a) We obtained a few ES-cell-like colonies from 5 × 105 DP31 cells transduced with OCT3/4 and SOX2 (OS), but we could not obtain any ES-cell-like colonies from DP75 cells. Human ES-cell-like colonies were counted at 30 days post infection. Error bars indicate ± S.D. (n = 3). (b) Genomic PCR using transgene-specific primers, with DP31 cells as a negative control, confirmed the insertion of only two transgenes in iPSCs derived from DP31 cells transduced with OS (iPS-DP31-OS) by PCR. The numbers denote different iPSC lines. We cropped the gels and blots for clarifying our presentation. The gels have been run under the same experimental conditions. (c) iPS-DP31-OS cells expressed pluripotency markers including SSEA3, TRA-1-60, TRA-1-81 and NANOG. Scale bar = 100 μm. (d) Pluripotency of iPS-DP31-OS cells was confirmed by EB-mediated differentiation and teratoma formation assay. Immunofluorescence staining showed that EB structures derived from iPS-DP31-OS cells expressed markers characteristic of the three germ layers including βIII-tubulin (ectoderm), α-smooth-muscle actin (mesoderm) and α-fetoprotein (endoderm). Nuclei were stained with Hoechst 33342. Scale bar = 100 μm. Hematoxylin- and eosin-stained sections of teratomas generated from iPS-DP31-OS cells are shown in the lower panels. The teratomas contained various tissues of all three germ layers, such as neural-tube-like structures (ectoderm), cartilage (mesoderm) and gut-like epithelial tissue (endoderm). Abbreviations: AFP, alpha-fetoprotein; α-SMA, alpha smooth muscle actin. Scale bar = 100 μm.
Statistical comparison of reprogramming potency between immature and mature tooth DPCs
To confirm that the reprogramming potency of DPC lines was dependent on the developmental stage but not on simple individual differences of donor teeth, we selected an additional five DPC lines (nine lines in total) isolated from various developmental stages (Supplementary Fig. S1) and introduced Yamanaka's four factors (OCT3/4, SOX2, KLF4, c-MYC; OSKM) into them using a retroviral gene-delivery system. Human ES-cell-like colonies were morphologically identified and counted at 21 days post-infection as previously described1,14. The number of human ES-cell-like colonies obtained from each DPC line was normalized to that of DP31 cells. All DPC lines isolated from the immature teeth (CC and RF stages) group showed significantly higher reprogramming efficiency than the lines isolated from mature (RC stage) teeth (Fig. 2a).
DPCs from immature teeth were more amenable to reprogramming than DPCs from mature teeth and showed high expression levels of endogenous DLX4.
(a) Nine DPC lines were selected for evaluation of reprogramming potency. The four Yamanaka factors (OSKM) were introduced by a retroviral system and the induced human ES-cell-like colonies were counted at 21 days post infection. The number of human ES-cell-like colonies obtained from each DPC line was normalized against that obtained from DP31 cells. Error bars indicate ± S.D. (n = 3). Asterisks indicate statistical significance: *P < 0.05, ** P < 0.01 and *** P < 0.001 compared to DP31 values. (b) The transcript levels of endogenous DLX4 were quantified using real-time RT-PCR. DPCs from immature teeth showed significantly higher levels of endogenous DLX4 than DPCs from mature teeth. Moreover, the transcript levels of DLX4 in all immature teeth DPCs were similar to those of human ES cells (hKES). GAPDH was used as an internal control. Error bars indicate ± S.D. (n = 3). Asterisks indicate statistical significance: *P < 0.05, ** P < 0.01 and *** P < 0.001 compared to values from DP31 cells.
Identification of differentially expressed genes in DPCs isolated from immature and mature teeth
The results of the reprogramming experiments prompted us to compare the gene expression profiles of DPCs from mature and immature teeth. To avoid masking of target genes by sex-related genes, we chose eight DPC lines isolated from male donors (four lines from individuals with immature teeth and four lines from individuals with mature teeth) (Supplementary Fig. S1). Among the 28 genes significantly up-regulated by more than five-fold in immature teeth DPCs (Supplementary Table S1), we focused on the homeobox gene Dlx4, the expression of which was up-regulated 35-fold in immature teeth DPCs on average and was thus the most differentially expressed transcription factor.
We performed a real-time PCR assay to evaluate the transcript levels of DLX4 in each DPC line. As expected, the transcript levels of DLX4 in DPCs from immature teeth were much higher than those of mature teeth DPCs and HDFs (Fig. 2b). Moreover, the transcript levels of DLX4 in all immature teeth DPCs were similar to those of human ES cells (hKES).
Ectopic expression of DLX4 promotes human iPSC generation
We next studied whether ectopic expression of DLX4 could increase the reprogramming efficiency of DPCs. Because we could not obtain any ES-cell-like colonies from mature teeth DPCs using the OS combination, we tried co-expression of DLX4 with OS (OSD) to convert DP75 into pluripotent cells. Contrary to our expectation, we could not obtain any ES-cell-like colonies from DP75 using the OSD cocktail (Fig. 3a). However, we observed a synergistic increase in the number of ES-cell-like colonies when both DLX4 and KLF4 were co-introduced with OS (OSKD) (~0.03%). The number of ES-cell-like colonies obtained from DP75 with OSKD was similar to the number obtained using OSKM (Fig. 3a). Next, we addressed whether ectopic expression of DLX4 could produce a similar effect during the reprogramming of DP31 cells obtained from immature teeth. As observed in DP75 cells, ectopic expression of DLX4 enhanced the reprogramming efficiency of DP31 cells (Fig. 3b). Although we could not obtain any ES-like colonies from 5 × 104 introduced DP31 cells with OS combination, we could obtain about 30 ES-like colonies constantly by using OSD combination. The OSD combination increased the reprogramming efficiency of DP31 (~0.07%) to the same level provided by OSK (Fig. 3b). As observed in DPCs, the OSKD cocktail increased the reprogramming efficiency in HDF cells to the same level observed using OSKM, although we did not obtain any ES-cell-like colonies with OSD alone, as observed for DP75 cells (Fig. 3c). Our results indicate that ectopic expression of DLX4 enhances reprogramming efficiency in combination with OCT3/4 and SOX2, similarly to KLF4. Notably, we found that use of the OSKD combination not only enhanced reprogramming, but also prevented the generation of non-ES-cell-like colonies to a greater extent than was observed with OSKM (Supplementary Fig. S2).
Ectopic expression of DLX4 promoted human iPSC generation.
(a-c) Numbers of human ES-cell-like colonies obtained from DPCs and HDFs. Human ES-cell-like colonies isolated from 5 × 105 or 5 × 104 infected cells were counted at 30 days post infection. Error bars indicate ± S.D. (n = 3). Asterisks indicate statistical significance; *P < 0.05, **P < 0.01 and ***P < 0.001.
To determine whether enhanced reprogramming efficiency by DLX4 overexpression could result in stable establishment of iPSC lines, we picked nine ES-cell-like colonies derived from DP31 with OSD (termed iPS-DP31-OSD) for further examination. Among them, eight colonies were successfully expanded. Immunostaining confirmed the expression of pluripotent markers, including OCT3/4, SSEA4, TRA-1-60 and TRA-1-81, at levels similar to those in human ES cells (Supplementary Fig. S3a). Using real-time PCR analysis, we found that transcript levels of the pluripotency markers OCT3/4 (endogenous), NANOG and REX1 were markedly increased in iPS-DP31-OSD lines relative to the parental cells and were similar to the transcript levels in human ES cells (Supplementary Fig. S3b). Differentiation potential towards the three germ layers was also confirmed in vitro and in vivo (Supplementary Fig. S3c).
Activation of TGF-β signal suppresses the expression level of DLX4 in DPCs and impairs iPSC generation from DPCs
Previous reports showed that MET (mesenchymal-to-epithelial transition) is a crucial early process for direct reprogramming of mouse fibroblasts to pluripotent status and that ectopic expression of KLF4 potently induced CDH1 (E-cadherin) mRNA to promote MET19,20,30. Similarly, KLF4 alone robustly induced CDH1 in DP31 cells as previously reported with mouse fibroblasts20,30 (Supplementary Fig. S4a). Interestingly, ectopic expression of DLX4 also up-regulated CDH1. However, the potency of CDH1 mRNA up-regulation appeared to be lower than that of KLF4. Recent reports showed that using an inhibitor of TGF-β signaling, which promotes EMT (epithelial to mesenchymal transition), could enhance iPSC generation from mouse and human fibroblasts20,31,32,33. We then attempted to address whether TGF-β signaling could affect nuclear reprogramming of DPCs. Interestingly, the expression level of endogenous DLX4 in DPCs was suppressed by TGF-β signaling (Fig. 4a). Moreover, addition of TGF-β1 dramatically reduced the number of iPSC colonies obtained from DP1 and DP31 cells (Fig. 4b). Our results indicated that activation of TGF-β signaling suppresses the expression of DLX4 in DPCs and impairs the generation of iPSC from DPCs.
Activation of TGF-β signal suppressed the expression level of DLX4 in DPCs and impaired the generation of iPSCs from DPCs.
(a) DP1 and DP31 cells were cultured in DPC culture medium with TGF-β1 (5 ng/mL) or with acetic acid (as a control) for 48 h. Real-time PCR analysis showed that after activation of TGF-β signal, the transcript levels of DLX4 were down-regulated in both DP1 and DP31 cells. GAPDH was used as an internal control. Error bars indicate ± S.D. (n = 3). Asterisks indicate statistical significance; ***P < 0.001 (b) Human ES-cell-like colonies isolated from 5 × 104 infected cells (OSKM) were counted at 21 days post infection. Numbers of human ES-cell-like colonies obtained from DP1 and DP31 were dramatically reduced when TGF-β1 (5 ng/mL) was added during the reprogramming process.
Since TGF-β signal also plays a crucial role in regulation of the cell cycle34,35 and acceleration of cell cycle is a crucial process in early stage of reprogramming21. Next, we addressed whether enforced expression of DLX4 could affect the cell cycle in DPCs. Firstly, we tested whether enforced expression of DLX4 can activate endogenous expression of c-MYC in DPCs, because of its potential to replace c-MYC during nuclear reprogramming. Contrary to our expectations, we could not observe the up-regulation of c-MYC in DLX4 induced DP31 cells (Supplementary Fig. S4b). Similar to this, we also could not observe the down-regulation of CDK inhibitors by ectopic expression of DLX4, even with the combination of OSK (Supplementary Fig. S4b). Our results indicated that DLX4 does not accelerate cell growth in the early stage of reprogramming to promote iPSC generation.
Discussion
In this study, we have identified a new transcription factor, DLX4, also known as DLX7 or BP1, which promotes human iPSC generation in combination with the classical Yamanaka factors. In mammals, the Dlx gene family consists of six members that are organized into three closely linked pairs: Dlx1/Dlx2, Dlx5/Dlx6 and Dlx3/Dlx436,37,38. These Dlx genes play crucial roles in early development39,40,41,42 and are known to be expressed in cranial neural crest cells and later in the craniofacial mesenchyme43. In this study, we found that DPCs isolated from developing teeth in the CC or RF stage show high expression of DLX4. Some reports have demonstrated that Dlx genes are associated with mammalian tooth formation39,44 and indicated that DLX4 is closely related to abnormal human tooth formation. For example, a mutation of DLX3, which is the partner of DLX4, is associated with tricho-dento-osseous (TDO) syndrome and disrupts odontoblast polarization and dentin formation45. Therefore, it is conceivable that DLX4 is highly expressed in DPCs isolated from third molar teeth, which are still under development even after birth.
Previously, Maekawa et al. attempted to identify new transcription factors with the ability to replace Klf4 during nuclear reprogramming of mouse skin fibroblasts from their library of 1,437 human transcription factors46. DLX4 was in their library, but was not included in the final screening. We could not obtain any ES-cell-like colonies from HDFs using the OSD cocktail. Conversely, we found that DLX4 promoted iPSC generation in HDFs only in the presence of KLF4. Therefore, it is possible that ectopic expression of DLX4 alone did not improve reprogramming efficiency in HDFs. It was thus excluded from the candidate list in the previous study.
Here we showed that the OS cocktail converted DP31 into a pluripotent state, although cells isolated from mature teeth (DP75) failed to be reprogrammed. This result indicates that DPCs isolated from immature teeth have unique properties. In this study, we have revealed that ectopic expression of DLX4 significantly promotes human iPSC generation; accordingly high expression levels of DLX4 in immature teeth DPCs are helpful for their effective reprogramming. Our results suggest that DLX4 plays a crucial role during reprogramming of DPCs and controls their reprogramming efficiency. However, the regulation of reprograming efficiency by DLX4 does not fully explain the unique properties of immature teeth DPCs. For example, the OSD cocktail was still insufficient to convert DP75 to a pluripotent state, conversely it up-regulated the reprogramming efficiency of DP31 to the same level provided by OSK. Moreover, DP87 shows substantially higher reprogramming efficiency but the expression level of DLX4 is average compared to other immature teeth DPC lines. This indicated that the measured endogenous DLX4 expression level determined by real-time PCR did not completely correlate with the reprogramming efficiency of DPCs and endogenous DLX4 expression may not be the sole factor responsible for the reprogramming efficiency to DPCs. A plausible explanation is that DLX4 may function together with some partner gene(s) to promote reprogramming, so the expression level of this (these) gene(s) may also affect the reprogramming efficiency of DPCs. Further screening of the genes listed in Supplemental Table S1 may lead to the discovery of additional factors that work together with DLX4.
An important remaining question is how DLX4 acts on a molecular level to promote reprogramming process in DPCs. Trinh et al. showed that DLX4 induces c-MYC and inhibits TGF-β mediated induction of p15Ink4B and p21WAF1/Cip1 expression by blocking the TGF-β/Smad signaling pathway in normal and malignant epithelial cell lines47. Blockage of TGF-β signaling leads to promotion of MET, which is critical initiating event during the derivation of iPSCs from mesenchymal cells19,20. In this study we also observed that enforced expression of DLX4 in DPCs caused modest up-regulation of CDH1, however compared to KLF4, the potential to induce CDH1 is very mild. It was initially speculated that DLX4 may promote reprogramming process by activating the endogenous expression of c-MYC, because we found that DLX4 can restore the reprogramming efficiencies of DPCs and HDFs in the absence of c-MYC. The close correlation between DLX4 and c-MYC also has been shown in other cell types, such as hematopoietic cells48,49. Contrary to our expectation, we could not observe the up-regulation of c-MYC after induction of DLX4 in DPCs. Similar to c-MYC, enforced expression of DLX4 also did not affect the transcript levels of CDK inhibitors, such as p21WAF1/Cip1, p15Ink4B and p16Ink4A, which have been demonstrated as roadblocks against reprogramming18,50,51,52,53. On the other hand, we did observe that TGF- β suppressed the expression of DLX4 in DPCs and impaired iPSC generation. Although the TGF-β/activin signaling pathway is important for the maintenance of human embryonic stem cells54,55, our results suggested that the TGF-β signaling associated with reduction of DLX4 is one of the roadblocks for effective reprogramming of human somatic cells. Since homeobox genes often function in a context dependent and lineage specific manner, the functions of DLX4 may be depended on the cell types. Although we have tested two mesenchymal linages of somatic cells whether DLX4 can promote reprogramming, it is desirable to test another cell types, such as epithelial and hematopoietic cells.
In this study we used conventional retroviral system to induce iPSCs and our process contains splitting step at 6 days post infection. So we cannot formally exclude the possibility that we might count the ES-like colonies derived from the same clones and this could result in overestimation of reprogramming efficiencies. But as mentioned above, it was unlikely that DLX4 induced proliferation in DPCs before the splitting step and accordingly the number of ES-like colonies obtained from using DLX4 was increased.
Here we show that DPCs isolated from immature teeth are more easily reprogrammed into iPSCs than DPCs derived from adult teeth and identify DLX4 as one of the candidate genes that is differentially expressed during the developmental stage. To our knowledge, this is the first report of a new transcript factor that enhances iPSC generation based on a comparative analysis of gene expression profiles using human cell resources. Our findings also provide evidence that our DPC collection obtained from many donors is a promising resource for both basic and clinical studies toward setting up a human iPS cell bank.
Methods
Cell culture
The isolation of DPCs was described in our previous report. DPCs were cultured in MSCGM (Lonza) and HDFs were cultured in DMEM with 10% FBS containing penicillin/streptomycin. The human ES cell line (KhES01) was obtained from Kyoto University and cultured on mitomycin C-treated SNL feeder cells in Primate ES cell medium (ReproCELL) supplemented with 4 ng/mL bFGF (WAKO). Human iPSCs were generated and maintained in the human ES cell culture condition.
Virus production and iPSC generation
We first introduced the mouse receptor for retroviruses (Slc7a1) into DPCs and HDFs using a lentiviral system according to our previous report1,14. Complementary DNA for human OCT3/4, SOX2, KLF4 and c-MYC was cloned into the retroviral pMXs vector and DLX4 was transferred to the pMXs-GW retroviral expression vector using the Gateway LR reaction. These pMXs-based retroviral vectors were separately transfected into Plat-E cells (ecotropic retrovirus packaging cells) using Fugene 6 (Roche). We also used a pMX retroviral vector encoding enhanced green fluorescent protein (EGFP) to monitor transduction efficiency by fluorescence-activated cell analysis (FACSAria, Beckton Dickinson). Twenty-four hours after transfection, the medium was replaced with fresh medium. After 24 hours, the viral supernatant was collected and mixed with an equal amount of each factor to infect HDFs/Slc7a1 and DPCs/Slc7a1 in medium supplemented with 4 μg/mL polybrene (Nacalai Tesque). Six days after retroviral infection, HDFs/Slc7a1 and DPCs/Slc7a1 were harvested by trypsinization and replated at 5 × 104 or 5 × 105 cells per 100-mm dish on mitomycin C-treated SNL feeder cells. The next day, the medium was replaced with primate ES cell medium supplemented with 4 ng/mL bFGF. Recombinant human TGF-β1 (reconstituted with 10 mM citric acid containing 0.1% BSA) was obtained from Pepro Tech. To evaluate the effect of activating TGF-β signal during reprogramming, we added TGF-β1 (5 ng/mL) into culture medium from day 1 to day 21 after retroviral infection and counted the human ES-cell-like colonies obtained from 5 × 104 infected DPCs at day 21.
Assessment of reprogramming efficiency of DPC lines
To determine the reprogramming potential of DPCs isolated from various developmental stages, we introduced the OSKM transcription factors into nine DPC lines using a retroviral system. The generation of iPSCs was performed according to our general protocol. Since we found that the transduction efficiencies of DPCs are at similar levels in their early passages, we used DPCs within ten passages to induce iPSCs. We counted the human ES-cell-like colonies obtained from 5 × 104 infected DPCs at 21 days after infection. The assays were conducted three times. The number of the human ES-cell-like colonies obtained from each DPC line was normalized to the number obtained from DP31. The relative reprogramming efficiency of DP31 was set as 1.0.
Real-time PCR
Total RNA was isolated from the cells using an RNeasy Plus Mini Kit (Qiagen). Total RNA (500 ng) was used for the reverse transcription reaction with Rever Tra Ace-α (Toyobo) according to the manufacturer's instructions. For real-time PCR analysis, PCR amplification of cDNA was performed by using SYBR Premix Ex Taq (Takara) and analyzed using a Thermal Cycler Dice Real-Time System (Takara). Primer sequences are listed in Supplementary Table S2.
Immunofluorescence staining
The cells were fixed with 4% paraformaldehyde and treated with PBS containing 5% normal goat or donkey serum, 1% BSA and 0.2% TritonX-100. The following antibodies were used: SSEA3 (1:10), TRA-1-81 (1:50), TRA-1-60 (1:50) (these antibodies were used at Kyoto University and were kind gifts from Dr. Peter W. Andrews), anti-SSEA4, anti-TRA-1-81, anti-TRA-1-60 (1:500, all contained in the ES Cell Characterization Kit from Merck Millipore; these antibodies were used at Gifu University), anti-NANOG (1:20, R&D Systems), anti-OCT3/4 (1:1000, Santa Cruz Biotechnology), anti-βIII-tubulin (1:200, Cell Signal Technology), anti-βIII-tubulin (1:2000, Covance), anti-α-SMA (1:500, DAKO) and anti-AFP (1:100, R&D). Nuclei were stained using 1 μg/mL Hoechst33342 (Life Technologies).
EB formation and differentiation assay
For EB formation, we harvested iPSCs by treating them with collagenase IV. The clumps of cells were transferred to poly (2-hydroxyethyl methacrylate)-coated dishes containing DMEM/F12 supplemented with 20% knockout serum replacement (Life Technologies), 2 mM L-glutamine, 0.1 M non-essential amino acids, 0.1 M 2-mercaptoethanol (Life Technologies) and 0.5% penicillin/streptomycin. The medium was changed every other day. After eight days as floating cultures, EBs were transferred to gelatin-coated plates and cultured in the same medium for another eight days. Spontaneous germ-layer differentiation of EBs was assayed using markers for endoderm (AFP), mesoderm (α-SMA) and ectoderm (βIII-tubulin).
Teratoma formation
For teratoma formation, 3 × 105 iPSCs were injected into the testes of a SCID mouse using a Hamilton syringe. Nine weeks after injection, tumors were dissected and fixed with PBS containing 4% paraformaldehyde. After paraffin embedding, the tissue was sectioned and stained with hematoxylin and eosin.
DNA microarray
Total RNA was prepared using the RNeasy Midi Kit (Qiagen). RNA (250 ng) was converted to cDNA, amplified and labeled with Cy3 using the Quick Amp Labeling Kit (Agilent). Labeled DNA was hybridized to a Whole Human Genome Microarray Kit, 4 × 44 K (Agilent) according to the manufacturer's protocol. After hybridization, the arrays were washed consecutively using a Gene Expression Wash Pack (Agilent) and scanned using the G2505C Microarray Scanner System (Agilent). The data were normalized and analyzed by using GeneSpringGX11 (Agilent). The level of gene expression was determined as the average difference and we selected genes with at least a five-fold difference in expression between DPCs from immature teeth and those from mature teeth. The microarray data was submitted to the NCBI GEO database under the accession number (GSE52853).
Statistical analysis
The differences in mean values were assessed by using unpaired Student's t-test, with P < 0.05 considered statistically significant. Error bars indicate ± SD (n = 3).
Additional Information
Accession codes: The microarray data were submitted to the NCBI GEO database under the accession number (GSE52853).
References
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007).
Aasen, T. et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 26, 1276–1284 (2008).
Aoi, T. et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 321, 699–702 (2008).
Eminli, S., Utikal, J., Arnold, K., Jaenisch, R. & Hochedlinger, K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells 26, 2467–2474 (2008).
Hanna, J. et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 133, 250–264 (2008).
Mali, P. et al. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells 26, 1998–2005 (2008).
Park, I. H. et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 (2008).
Stadtfeld, M., Brennand, K. & Hochedlinger, K. Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Curr. Biol. 18, 890–894 (2008).
Haase, A. et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell 5, 434–441 (2009).
Loh, Y. H. et al. Generation of induced pluripotent stem cells from human blood. Blood 113, 5476–5479 (2009).
Sun, N. et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc. Natl. Acad. Sci. USA 106, 15720–15725 (2009).
Utikal, J., Maherali, N., Kulalert, W. & Hochedlinger, K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J. Cell Sci. 122, 3502–3510 (2009).
Tamaoki, N. et al. Dental pulp cells for induced pluripotent stem cell banking. J. Dent. Res. 89, 773–778 (2010).
Giorgetti, A. et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell 5, 353–357 (2009).
Kim, J. B. et al. Direct reprogramming of human neural stem cells by OCT4. Nature 461, 649–643 (2009).
Stadtfeld, M., Maherali, N., Breault, D. T. & Hochedlinger, K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2, 230–240 (2008).
Banito, A. et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 23, 2134–2139 (2009).
Chen, T. et al. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells 28, 1315–1325 (2010).
Li, R. et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 (2010).
Ruiz, S. et al. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Current Biol.: CB 21, 45–52 (2011).
Kim, J. B. et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454, 646–650 (2008).
Eminli, S. et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat. Genetics 41, 968–976 (2009).
Kim, J. B. et al. Oct4-induced pluripotency in adult neural stem cells. Cell 136, 411–419 (2009).
Gronthos, S., Mankani, M., Brahim, J., Robey, P. G. & Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 97, 13625–13630 (2000).
Gronthos, S. et al. Stem cell properties of human dental pulp stem cells. J Dent Res 81, 531–535 (2002).
Stevens, A. et al. Human dental pulp stem cells differentiate into neural crest-derived melanocytes and have label-retaining and sphere-forming abilities. Stem Cells Dev. 17, 1175–1184 (2008).
Takeda, T. et al. Characterization of dental pulp stem cells of human tooth germs. J. Dent. Res. 87, 676–681 (2008).
Zhang, W. et al. In vivo evaluation of human dental pulp stem cells differentiated towards multiple lineages. J. Tissue Eng. Regen. Med. 2, 117–125 (2008).
Chen, J. et al. BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell Res. 21, 205–212 (2011).
Ichida, J. K. et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 5, 491–503 (2009).
Lin, T. et al. A chemical platform for improved induction of human iPSCs. Nat. Methods 6, 805–808 (2009).
Maherali, N. & Hochedlinger, K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol: CB 19, 1718–1723 (2009).
Datto, M. B., Yu, Y. & Wang, X. F. Functional analysis of the transforming growth factor beta responsive elements in the WAF1/Cip1/p21 promoter. J. Biol. Chem. 270, 28623–28628 (1995).
Reynisdottir, I., Polyak, K., Iavarone, A. & Massague, J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev 9, 1831–1845 (1995).
Merlo, G. R. et al. Multiple functions of Dlx genes. Int. J. Dev. Biol. 44, 619–626 (2000).
Panganiban, G. & Rubenstein, J. L. Developmental functions of the Distal-less/Dlx homeobox genes. Development 129, 4371–4386 (2002).
Morasso, M. I. & Radoja, N. Dlx genes, p63 and ectodermal dysplasias. Birth Defects Res. Part C, Embryo Today: Rev. 75, 163–171 (2005).
Thomas, B. L. et al. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development 124, 4811–4818 (1997).
Depew, M. J., Lufkin, T. & Rubenstein, J. L. Specification of jaw subdivisions by Dlx genes. Science 298, 381–385 (2002).
Robledo, R. F., Rajan, L., Li, X. & Lufkin, T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial and appendicular skeletal development. Genes Dev. 16, 1089–1101 (2002).
Heude, E. et al. Jaw muscularization requires Dlx expression by cranial neural crest cells. Proc. Natl. Acad. Sci. USA 107, 11441–11446 (2010).
Chai, Y. et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127, 1671–1679 (2000).
Duverger, O. et al. Neural crest deletion of Dlx3 leads to major dentin defects through down-regulation of Dspp. J. Biol. Chem. 287, 12230–12240 (2012).
Price, J. A., Bowden, D. W., Wright, J. T., Pettenati, M. J. & Hart, T. C. Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Human Mol. Genetics 7, 563–569 (1998).
Maekawa, M. et al. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 474, 225–229 (2011).
Trinh, B. Q., Barengo, N. & Naora, H. Homeodomain protein DLX4 counteracts key transcriptional control mechanisms of the TGF-beta cytostatic program and blocks the antiproliferative effect of TGF-beta. Oncogene 30, 2718–2729 (2011).
Shimamoto, T., Nakamura, S., Bollekens, J., Ruddle, F. H. & Takeshita, K. Inhibition of DLX-7 homeobox gene causes decreased expression of GATA-1 and c-myc genes and apoptosis. Proc. Natl. Acad. Sci. USA 94, 3245–3249 (1997).
Awwad, R. T. et al. Overexpression of BP1, a homeobox gene, is associated with resistance to all-trans retinoic acid in acute promyelocytic leukemia cells. Ann Hematol 87, 195–203 (2008).
Hong, H. et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460, 1132–1135 (2009).
Li, H. et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460, 1136–1139 (2009).
Utikal, J. et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460, 1145–1148 (2009).
Xu, Y. et al. Proliferation rate of somatic cells affects reprogramming efficiency. J. Biol. Chem. 288, 9767–9778 (2013).
Jiang, J. & Ng, H. H. TGFbeta and SMADs talk to NANOG in human embryonic stem cells. Cell stem cell 3, 127–128 (2008).
Xu, R. H. et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell stem cell 3, 196–206 (2008).
Acknowledgements
We thank Y. Kawamura (AIST) for technical support for the DLX4 expression vector, T. Tanabe (CiRA) for technical support for the OS-induced iPSCs and T. Ichisaka (CiRA) for technical support of teratoma assay. We also thank our laboratory members for their technical help and fruitful discussions. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the JST Yamanaka iPS Cell Project.
Author information
Authors and Affiliations
Contributions
N.T. performed and designed most of the experiments, analyzed the data and wrote the manuscript. K.T. performed and designed the experiments for OS-induced iPSCs. H.A. and M.I. performed the characterization of iPSCs. K.I. and D.H. generated iPSCs from DPCs. T.K. performed the computer analyses of the DNA microarray data. N.C., A.I., T.K. and T.S. analyzed all data in this study. N.G. provided the retroviral expression clones. S.Y. supervised this study. K.T. conceived and supervised this study, designed the experiments, analyzed the data and wrote and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Tamaoki, N., Takahashi, K., Aoki, H. et al. The homeobox gene DLX4 promotes generation of human induced pluripotent stem cells. Sci Rep 4, 7283 (2014). https://doi.org/10.1038/srep07283
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07283
This article is cited by
-
METTL3-mediated m6A modification regulates cell cycle progression of dental pulp stem cells
Stem Cell Research & Therapy (2021)
-
FGF2-responsive genes in human dental pulp cells assessed using a rat spinal cord injury model
Journal of Bone and Mineral Metabolism (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.