Abstract
Ztbt20 is a POK family transcription factor and primarily functions through its conserved C2H2 Krüppel type zinc finger and BTB/POZ domains. The present study was designed to define the function of the Zbtb20, in vivo, during mouse spermatogenesis. Immunohistochemical studies revealed that ZBTB20 protein was localized specifically in the nuclei of Sertoli cells in seminiferous tubules. To investigate its role during spermatogenesis, we crossed Amh-Cre transgenic mice with Zbtb20 floxp mice to generate conditionally knockout mice (cKO) in which Zbtb20 was specifically deleted in Sertoli cells. The cKO mice were fertile and did not show any detectable abnormalities in spermatogenesis. Taken together, though specific deletion of transcription factor Zbtb20 in Sertoli cells has no apparent influence on spermatogenesis, its specific localization in Sertoli cells makes Zbtb20 a useful marker for the identification of Sertoli cells in seminiferous tubules.
Similar content being viewed by others
Introduction
Transcription factors are regulatory proteins that are mainly involved in transcription activation (or rarely in inhibition) by binding to DNA sequences1,2. They are considered to be the most important regulators in gene expression2,3. Transcription factors work in germ and somatic cells (especially in Sertoli cells) to control spermatogenesis in mammals4,5. So far, only relatively few transcription factors have been identified in spermatogenesis. Studies of all the transcription factors functioning in spermatogenesis indicate that transcription factors expressing in germ cells are more likely to function in specifically developmental stages of germ cells, while transcription factors expressing in Sertoli cells are often required during all the stages of gametogenesis in seminiferous tubules4,5. ZBTB [zinc finger and BTB (Broad complex, Tramtrack and Bric-à-brac)] proteins, also known as Poxvirus and zinc finger (POZ) and Krüppel-type (POK) proteins, are a family of transcription factors that play critical roles in development, differentiation and oncogenesis6,7,8,9,10,11,12. POK/ZBTB proteins exhibit C-terminal zinc fingers and an N-terminal BTB/POZ domain. The former recognizes and binds to specific DNA sequences, while the BTB/POZ domain mediates homodimerization and/or heterodimerization and interacts with other proteins13,14,15. In the mouse genome, 44 genes encode different POK/ZBTB proteins and a few of them have been reported to be essential for spermatogenesis. For example, Zbtb16, also known as Plzf (promyelocytic leukemia zinc finger), expresses in gonocytes and undifferentiated spermatogonia and it is the first gene reported to be essential for spermatogonial stem cell self-renewal in mouse testis9,16.
Besides Zbtb16, functional roles of other POK/ZBTB genes, such as Zbtb20, remain unclear during spermatogenesis. Zbtb20 overexpression in transgenic mice under the control of a forebrain-specific promoter (D6 promoter) leads to the formation of hippocampus-like neuronal structures and behavioral abnormalities17, which indicates that Zbtb20 can control the organ development and differentiation. Another study shows that Zbtb20 null mice were infertile, however, the underlying mechanisms remained unknown due to pleiotropic defects such as growth retardation, prolonged hypoglycemia and postnatal lethality before 12 weeks of age of these mice18.
The aim of this study was to define the function of the Zbtb20 in vivo during spermatogenesis of mice. We found that ZBTB20 specifically expressed in the nuclei of Sertoli cells, making it a useful marker for the identification of Sertoli cells in seminiferous tubes. However, mice homozygous for a null mutation of Zbtb20, specifically in Sertoli cells, do not exhibit any detectable abnormalities in spermatogenesis and fertility, indicating that Zbtb20 is dispensable for spermatogenesis and male fertility in mice.
Methods
Experimental animals
Zbtb20fl/fl mice homozygous for a floxp allele of Zbtb20, Amh-Cre mice and mT/mG reporter mice were used in the present study and were described previously19,20,21. For fertility testing, 8 to 12-week old Zbtb20fl/fl (control) and Amh-Cre; Zbtb20fl/fl (cKO) males were separately housed with wild-type C57BL/6 females for 6 months. The size of litters generated by these males was recorded. All mice were housed under controlled photoperiod conditions (lights on 08:00–20:00) and supplied with food and ddH2O ad libitum. All experimental protocols and animal handling procedures were conducted in accordance with the guidelines and procedures approved by Institutional Animal Care Committee of University of Science and Technology of China.
Sertoli cell isolation
A modified previously described method was used to isolate Sertoli cells from the testes of 21-dpp mice22,23. Briefly, testes were decapsulated under a dissection microscope. The seminiferous tubules were pooled and washed with phosphate-buffered saline (PBS) thrice and incubated with 2 mg/ml collagenase I (Sigma, C7661, MO, USA) and 0.5 mg/ml DNase I (Sigma, D4527) in DMEM: F12 (HyClone, SH40007-13, UT, USA) for 15 minutes at 37°C on a shaker, then washed twice with DMEM: F12 and further digested with 2 mg/ml collagenase I, 0.5 mg/ml DNase I and 1 mg/ml hyaluronidase (Sigma, H3506) for 15 minutes at 37°C. The tubules were allowed to settle and were then washed twice with DMEM: F12 before being digested with 2 mg/ml collagenase I, 0.5 mg/ml DNase I, 2 mg/ml hyaluronidase and 1 mg/ml trypsin (Sigma, T8003) for 30 minutes at 37°C. These dispersed cells were then washed twice with DMEM: F12 medium and placed into culture dishes in DMEM: F12 containing 10% fetal bovine serum (HyClone, SV30087-02) and were incubated at 37°C with 5% CO2. After 1 day of culturing, the solution was removed and the cells were treated with a hypotonic solution (20 mM Tris, pH 7.4) for 1 minutes for the removal of remaining germ cells, if any and harvested for further analyses.
Sperm counting
The unilateral epididymides and vasa deferentia were removed from 70-dpp control and cKO mice, incised several times and incubated in 1 ml buffer containing 75 mM NaCl, 24 mM EDTA and 0.4% bovine serum albumin (Sigma, A2058) at 37°C with 5% CO2 for 30 minutes to allow sperm release from the epididymides. Sperm were collected after a nylon-mesh filtration and counted with a haemocytometer.
Histological examination and immunohistochemistry
The control and cKO mice were euthanized by cervical dislocation and testes were immediately fixed in Bouin's solution for hematoxylin and eosin (H & E) staining or in 4% formaldehyde in PBS for immunohistochemistry. For immunohistochemistry, anti-SOX9 (1:100; Millipore, AB5535, MA, USA), anti-3β-HSD (1:100; Santa Cruz, SC-30820, CA, USA), anti-α-ACTIN2 (1:100; Abcam, AB68167) or anti-DDX4 (1:100; Abcam, AB13840, Cambridge, UK) were co-incubated with anti-ZBTB20 (1:500, donated by Prof. Weiping Zhang) respectively and the ZBTB20 signals were detected using a tyramine amplification kit (Invitrogen, T20948, MA, USA) according to the manufacturer's instructions. All images were captured using a microscope (Nikon Eclipse 80i microscope) equipped with a digital camera (Nikon DS-Ri1 for H & E staining or Hamamatsu C4742-80 for immunohistochemistry).
Real-time PCR
RNA isolation and real-time PCR were performed as previously described24. All PCR primers used are listed in Supplementary Table 1. For real-time PCR analysis, CT values of samples were normalized to the corresponding CT values of Gapdh. Quantification of the fold change in gene expression was determined by the comparative CT method.
Western blot
Protein samples from Sertoli cells were prepared in lysis buffer (50 mM Tris/HCl pH 7.4, 300 mM NaCl, 5 mM EDTA, 1% Triton X-100) supplemented with protease inhibitors (Roche). Western blot was carried out as described previously24 and primary antibodies against ZBTB20 (1:500) and GAPDH (1:1000; Millipore, MAB374, MA, USA) were used to detect the protein bands.
Statistical analysis
Protein and mRNA levels, testis weights, testis/body weight ratios, sperm number and litter size between control and cKO mice were analyzed by using the Student's t-test. Results are presented as mean ± SEM. Statistical significance was set at P < 0.05.
Results
Expression of ZBTB20 in mouse testes
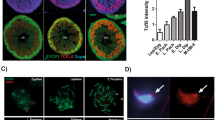
In seminiferous tubules of adult testes (70-dpp) (Fig. 1A), ZBTB20 protein was detected only in the nuclei of Sertoli cells, as it was co-localized with SOX9, a Sertoli cell nucleus specific marker (Fig. 1A) and it was not detected in germ cells as ZBTB20 staining was absent in the cells stained positive by DDX4, a germ cell specific marker (Fig. 1B). In order to more precisely evaluate ZBTB20 expression in Sertoli cells during testicular development, we performed immunohistological analysis of ZBTB20 in testes during early developmental stages (at 7- and 14-dpp) and observed that ZBTB20 was expressed at all the studied stages in Sertoli cells (Fig. 1A). Besides in Sertoli cells, ZBTB20 was also found in interstitial regions between seminiferous tubules in testis (Fig. 1A). Co-immunostaining of ZBTB20 with 3β-HSD, a Leydig cell cytoplasm specific marker, revealed that ZBTB20 was localized in Leydig cells (Supplementary Fig. S1A). Additionally, co-immunostaining of ZBTB20 with α-ACTIN2, a peritubular myoid cell marker, indicated that ZBTB20 also localized in some peritubular myoid cells (Supplementary Fig. S1B).
ZBTB20 protein localization in mouse testes.
(A). Cellular localization of ZBTB20 and SOX9 in testes at indicated ages. Localization of ZBTB20 (green) and SOX9 (red, a Sertoli cell marker) in sections of testes from 7-, 14- and 70-dpp mice was displayed by co-immunofluorescent staining for these proteins, showing that in seminiferous tubules, ZBTB20 just localized in the nuclei of Sertoli cells of all developmental stages examined. The images shown are representative results of experiments that were repeated thrice using samples from different sets of testes and yielded similar results. Scale bars, 50 μm. (B). Cellular localization of ZBTB20 and DDX4 in adult testes. Localization of ZBTB20 (green) and DDX4 (red, a germ cell marker) was observed in sections of testes from 70-dpp mice by immunofluorescence. In seminiferous tubules, opposite staining of ZBTB20 and DDX4 revealed that ZBTB20 did not localize in germ cells. The data shown are representative images of experiments that were repeated thrice using samples from different sets of testes and yielded similar results. Scale bars, 50 μm.
Zbtb20 deletion in Sertoli cells
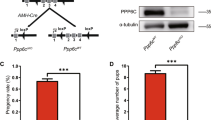
In order to assess the Zbtb20 functions in Sertoli cells during spermatogenesis, we generated mice in which the Zbtb20 gene was specifically disrupted in testicular Sertoli cells. We combined a conditional floxp Zbtb20 allele with the Cre-expressing line Amh-Cre (Fig. 2A). To study the CRE recombinase activity, Amh-cre mice were crossed with fluorescence reporter mice (mT/mG), in which Amh-Cre mediated recombination resulted in excision of the RFP cassette and expression of the GFP reporter21. Based on the analysis of Amh-Cre recombined mT/mG reporter mice, we confirmed that the Amh-Cre mice had strong recombinase activities in Sertoli cells (Supplementary Fig. S2). Zbtb20 deletion efficiency in Sertoli cells was analyzed by detecting the Zbtb20 mRNA and protein levels in the isolated Sertoli cells (Supplementary Fig. S3). We found that the Zbtb20 disruption resulted in a drastic reduction in Zbtb20 mRNA and protein levels in the cKO Sertoli cells (Supplementary Fig. S4 and Fig. 2B–C). As shown in Fig. 2D, immunofluorescent analysis of the cKO testes revealed that the localization of ZBTB20 in Sertoli cell was abolished (Fig. 2D) without affecting its localization in Leydig cells (Supplementary Fig. S5), which further confirms the efficient and specific disruption of Zbtb20 in Sertoli cells.
Conditional deletion of Zbtb20 in Sertoli cells.
(A). Hybrid scheme used to develop Zbtb20 cKO mice. (B). Western blot analysis of ZBTB20 in isolated Sertoli cells from 21-dpp control and cKO mice. GAPDH is served as a protein loading control. The image shown is a representative result from three independent experiments. (C). Quantitative results of Western blot experiments shown in (B). The ZBTB20 protein level was normalized and plotted against GAPDH and was arbitrarily set as 1 in controls. Data are presented as mean ± SEM from three independent experiments. **P < 0.01, Student's t-test. (D). Cellular localization of ZBTB20 and SOX9 in sections of control and cKO testes. Localization of ZBTB20 (green) and SOX9 (red) in sections of testes of 21-dpp mice were shown by immunofluorescence. ZBTB20 localization was lost in cKO Sertoli cells. The shown are representative images from experiments that were repeated thrice using samples from different sets of testes and yielded similar results. Scale bars, 50 μm.
Normal spermatogenesis in Zbtb20 cKO mice
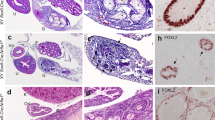
After successfully generating a Sertoli cell Zbtb20 knockout mouse model, we investigated spermatogenesis in the cKO mice. The testes from control and cKO littermates of different ages were analysed. We did not find any significant differences in testis morphology, weight and testis/body weight ratio between the two groups (Fig. 3A–B; Supplementary Fig. S6). As shown in Fig. 3C, both cKO and control testes exhibited typical seminiferous tubule morphology with all stages of spermatogenic cells (from spermatogonia to spermatozoa), suggesting that spermatogenesis was normal in cKO mice. We also analyzed the sperm in epididymides and found that cKO and control mice produced similar number of sperm (Fig. 3C–D). Finally, the breeding performance of the cKO males was found similar to that of control males as litters produced by mating cKO males with wild type females showed a size similar to those produced by control males (Fig. 3E). Taken together, these results indicated that Zbtb20 conditional knockout in Sertoli cells does not affect spermatogenesis or fertility of mice.
Normal spermatogenesis in cKO mouse testes.
(A). Representative images of testes from 70-dpp control and cKO mice. Scale bars, 2 mm. (B). Mean testis weight of 70-dpp control and cKO mice. n, the number of animals. Error bars represent SEM. (C). H&E staining of testes and epididymides from control and cKO mice at indicated ages. a–h: Testes from control and cKO mice of age at 21-(a, b), 42-(c, d), 70-(e, f) and 140-(g, h) dpp. i–l: Epididymides caput (i, j) and cauda (k, l) from 70-dpp control (i, k) and cKO mice (j, l). Scale bars, 50 μm. (D). Total sperm per epididymis from 70-dpp control and cKO mice. n, the number of animals. Error bars represent SEM. (E). The number of pups per litter from control and cKO mice. Data shown are from 6 control or 5 cKO males. Dots and squares represent the number of pups of each litter for control and cKO mice, respectively. n, the number of litters. Error bars represent SEM.
Discussion
Our present study describes the expression of transcription factor Zbtb20 in mouse testis. We have observed that ZBTB20 protein was specifically localized in the nuclei of Sertoli cells locating within seminiferous tubules along with Leydig and some peritubular myoid cells present between seminiferous tubes in mouse testes. As compared to its paralog gene, Zbtb16, which is restricted to gonocytes and undifferentiated spermatogonia9,16, Zbtb20 was specifically expressed in testicular somatic cells (Fig. 1A–B; Supplementary Fig. S1). In the interstitial compartment Leydig and some peritubular myoid cells showed a positive ZBTB20 staining (Supplementary Fig. S1). In contrast, all Sertoli cells showed a constant and intense expression of Zbtb20 in all seminiferous tubules independent of the age of animals, which makes it a useful molecular marker for the identification of Sertoli cells (Fig. 1A–B). This expression pattern was in agreement with the expression of previously described Sertoli cell transcriptional factors, such as SOX9 and GATA4, which have been shown to play essential roles in spermatogenesis25,26,27,28,29,30,31,32.
To define the function of Zbtb20 in Sertoli cells, we generated mice with the Zbtb20 being specifically disrupted in these cells by crossing Amh-Cre transgenic mice with Zbtb20 floxp mice (Fig. 2A). Note that, as the Amh-Cre transgene is expressed from embryonic day 14.5 onwards20, excision of Zbtb20 occurs long before the onset of spermatogenesis at 5-dpp33. Consistently, based on the Amh-Cre recombined mT/mG reporter mice, we confirmed that Amh-Cre mice had strong recombinase activity in Sertoli cells (Supplementary Fig. S2). In the isolated Sertoli cells, we observed that the Zbtb20 deletion using Amh-Cre resulted in a drastic reduction of Zbtb20 mRNA and protein level in the Sertoli cells of cKO mice (Fig. 2B–C; Supplementary Fig. S3–4). Furthermore, the Sertoli cell expression pattern of ZBTB20 was abolished in the cKO testes based on immunofluorescence analysis, further confirming efficient deletion of Zbtb20 in these cells (Fig. 2D and Supplementary Fig. S5). However, the phenotype of the Zbtb20 deficient mice indicates that Zbtb20 in Sertoli cells was not required for spermatogenesis (Fig. 3A–E; Supplementary Fig. S6).
The lack of phenotype in mice that Zbtb20 was specifically disrupted in Sertoli cells may result from functional redundancy in the Zbtb family. In this scenario, expression of Zbtb20 target genes in Zbtb20 null Sertoli cells would be regulated by other Zbtb factors, such as Zbtb32, Zbtb37 and Zbtb44, which are highly expressed in testes34. We thus hypothesize that some of the unidentified ZBTB proteins may have compensated for the loss of Zbtb20 function in Sertoli cells. ZBTB20 was also expressed in Leydig and peritubular myoid cells, however, the significance of this protein in these cells remains unclear. Thus, future studies on Leydig- or peritubular myoid cell-specific knockout animals for Zbtb20 may further shed light on the role of this transcription factor in the testes.
References
Latchman, D. S. Transcription factors: an overview. Int J Biochem Cell Biol 29, 1305–12 (1997).
Lee, T. I. & Young, R. A. Transcription of eukaryotic protein-coding genes. Annu Rev Genet 34, 77–137 (2000).
Mitchell, P. J. & Tjian, R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245, 371–8 (1989).
Bettegowda, A. & Wilkinson, M. F. Transcription and post-transcriptional regulation of spermatogenesis. Philos Trans R Soc Lond B Biol Sci 365, 1637–51 (2010).
Kanatsu-Shinohara, M. & Shinohara, T. Spermatogonial stem cell self-renewal and development. Annu Rev Cell Dev Biol 29, 163–87 (2013).
Lee, S. U. & Maeda, T. POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol Rev 247, 107–19 (2012).
Barna, M., Hawe, N., Niswander, L. & Pandolfi, P. P. Plzf regulates limb and axial skeletal patterning. Nat Genet 25, 166–72 (2000).
He, X. et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–33 (2005).
Costoya, J. A. et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 36, 653–9 (2004).
Ye, B. H. et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet 16, 161–70 (1997).
Koken, M. H. et al. Leukemia-associated retinoic acid receptor alpha fusion partners, PML and PLZF, heterodimerize and colocalize to nuclear bodies. Proc Natl Acad Sci U S A 94, 10255–60 (1997).
Maeda, T. et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature 433, 278–85 (2005).
Stogios, P. J., Downs, G. S., Jauhal, J. J., Nandra, S. K. & Prive, G. G. Sequence and structural analysis of BTB domain proteins. Genome Biol 6, R82 (2005).
Chang, C. C., Ye, B. H., Chaganti, R. S. & Dalla-Favera, R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci U S A 93, 6947–52 (1996).
Dhordain, P. et al. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci U S A 94, 10762–7 (1997).
Buaas, F. W. et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 36, 647–52 (2004).
Nielsen, J. V., Nielsen, F. H., Ismail, R., Noraberg, J. & Jensen, N. A. Hippocampus-like corticoneurogenesis induced by two isoforms of the BTB-zinc finger gene Zbtb20 in mice. Development 134, 1133–40 (2007).
Sutherland, A. P. et al. Zinc finger protein Zbtb20 is essential for postnatal survival and glucose homeostasis. Mol Cell Biol 29, 2804–15 (2009).
Xie, Z. et al. Zinc finger protein ZBTB20 is a key repressor of alpha-fetoprotein gene transcription in liver. Proc Natl Acad Sci U S A 105, 10859–64 (2008).
Lecureuil, C., Fontaine, I., Crepieux, P. & Guillou, F. Sertoli and granulosa cell-specific Cre recombinase activity in transgenic mice. Genesis 33, 114–8 (2002).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
van der Wee, K. S., Johnson, E. W., Dirami, G., Dym, T. M. & Hofmann, M. C. Immunomagnetic isolation and long-term culture of mouse type A spermatogonia. J Androl 22, 696–704 (2001).
Chang, Y. F., Lee-Chang, J. S., Panneerdoss, S., MacLean, J. A., 2nd & Rao, M. K. Isolation of Sertoli, Leydig and spermatogenic cells from the mouse testis. Biotechniques 51, 341–2, 344 (2011).
Zhang, H. et al. microRNA 376a regulates follicle assembly by targeting Pcna in fetal and neonatal mouse ovaries. Reproduction 148, 43–54 (2014).
Morais da Silva, S. et al. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat Genet 14, 62–8 (1996).
Chaboissier, M. C. et al. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 131, 1891–901 (2004).
Bishop, C. E. et al. A transgenic insertion upstream of sox9 is associated with dominant XX sex reversal in the mouse. Nat Genet 26, 490–4 (2000).
Jeske, Y. W., Bowles, J., Greenfield, A. & Koopman, P. Expression of a linear Sry transcript in the mouse genital ridge. Nat Genet 10, 480–2 (1995).
Kobayashi, A., Chang, H., Chaboissier, M. C., Schedl, A. & Behringer, R. R. Sox9 in testis determination. Ann N Y Acad Sci 1061, 9–17 (2005).
Ketola, I. et al. Expression and regulation of transcription factors GATA-4 and GATA-6 in developing mouse testis. Endocrinology 140, 1470–80 (1999).
Lavoie, H. A., McCoy, G. L. & Blake, C. A. Expression of the GATA-4 and GATA-6 transcription factors in the fetal rat gonad and in the ovary during postnatal development and pregnancy. Mol Cell Endocrinol 227, 31–40 (2004).
Kyronlahti, A. et al. GATA4 regulates Sertoli cell function and fertility in adult male mice. Mol Cell Endocrinol 333, 85–95 (2011).
Bellve, A. R. et al. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol 74, 68–85 (1977).
Shima, J. E., McLean, D. J., McCarrey, J. R. & Griswold, M. D. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 71, 319–30 (2004).
Acknowledgements
We thank Dr. Weiping Zhang (Department of Pathophysiology, Second Military Medical University, Shanghai, China) for his generous donation of Zbtb20 floxp mice and ZBTB20.This work was supported by the National Basic Research Program (2013CB947900 and 2014CB943101) of China (973), by grants from National Natural Science Foundation of China (31371519, 313111245 and 31301227) and the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-R-07).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: Q.S., X.J., H.Z. and W.Y. Performed the experiments: X.J., H.Z., S.Y., Y.Z., W.Y., W.Z., L.W. and Z.W. Analyzed the data: X.J., S.Y. and W.Y. Paper wrote up: Q.S. and X.J. Modification of the manuscript: I.B. F.I. and H.J.C.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Data
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Jiang, X., Zhang, H., Yin, S. et al. Specific deficiency of Plzf paralog, Zbtb20, in Sertoli cells does not affect spermatogenesis and fertility in mice. Sci Rep 4, 7062 (2014). https://doi.org/10.1038/srep07062
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07062
This article is cited by
-
Truncating PICK1 Variant Identified in Azoospermia Affected Mitochondrial Dysfunction in Knockout Mice
Current Medical Science (2023)
-
The evolutionarily conserved genes: Tex37, Ccdc73, Prss55 and Nxt2 are dispensable for fertility in mice
Scientific Reports (2018)
-
RNAi as a tool to control the sex ratio of mouse offspring by interrupting Zfx/Zfy genes in the testis
Mammalian Genome (2017)
-
Upregulation of PLZF is Associated with Neuronal Injury in Lipopolysaccharide-Induced Neuroinflammation
Neurochemical Research (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.