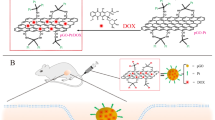
Abstract
Graphene quantum dots (GQDs) maintain the intrinsic layered structural motif of graphene but with smaller lateral size and abundant periphery carboxylic groups and are more compatible with biological system, thus are promising nanomaterials for therapeutic applications. Here we show that GQDs have a superb ability in drug delivery and anti-cancer activity boost without any pre-modification due to their unique structural properties. They could efficiently deliver doxorubicin (DOX) to the nucleus through DOX/GQD conjugates, because the conjugates assume different cellular and nuclear internalization pathways comparing to free DOX. Also, the conjugates could enhance DNA cleavage activity of DOX markedly. This enhancement combining with efficient nuclear delivery improved cytotoxicity of DOX dramatically. Furthermore, the DOX/GQD conjugates could also increase the nuclear uptake and cytotoxicity of DOX to drug-resistant cancer cells indicating that the conjugates may be capable to increase chemotherapy efficacy of anti-cancer drugs that are suboptimal due to the drug resistance.
Similar content being viewed by others
Introduction
Nanomaterials, due to their unique nanoarchitectures and diverse properties offer an unprecedented opportunity for improving drug loading, targeting and efficacy1,2. To this end, various nanostructured materials, such as carbon nanotubes3, polymeric nanoconjugates4,5, nanoparticles etc.6,7 have been explored, especially in cancer therapy. However, the delivery of the anti-cancer drugs to the nucleus by nanostructured materials remains a significant challenge8,9,10,11. Engineering the surfaces of nanomaterials with nuclear-targeted peptide, or chemicals could enhance the nuclear uptake and cytotoxicity of the drugs, but the modification dramatically increases the complexity in vivo, such as opsonization and difficulty for preparation and application12,13,14.
Recently, there has been significant biomedical research interest in graphene and graphene oxide (GO) because of their single atomic-layered structure and chemical properties15,16,17. Their therapeutic applications harnessed the opportunities that provided by graphene and GO to improve the drugs solubility18, extend their half-life19,20 and to reduce their side effects21,22. For instances, Dai et al pioneered the research in this area that PEG modified GO could increase the aqueous solubility of the anti-cancer drugs23. Zhang et al modified nano-scaled GO with folic acid (FA) then loaded the mixed anti-cancer drugs to specifically target the cells with FA receptors24. In most work so far reported, the drug loading on the pre-modified graphene or GO sheets was through hydrophobic and π-π stacking, but the release of anti-cancer drug was pH responsive21,22. And in most cases, the cytotoxicity was increased by the graphene or GO based delivery systems, but there is no experimental evidence showing that the accumulation of the anti-cancer drugs in the nucleus was improved, which makes their functional mechanisms are unclear. Moreover, graphene or GO sheets have been almost exclusively employed as drug carriers using their single atomic-layered structural feature, the intrinsic biological function of graphene, GO or their derivatives is ignored.
In our previous work, we found that GO sheets combining with copper ions could cleave the plasmid DNA into nicked DNA suggesting that GO sheets could be potentially useful in anti-cancer therapy25. Inspired by that finding, we further explored the impact of the GO on the small chemical nucleases and found that the nuclease activity and cytotoxicity of the chemical nucleases were both enhanced markedly upon combining with GO sheets26. These findings and other complimentary results lead us to believe that a conjugate of GO/chemical nucleases results in an improved nuclease activity and cytotoxicity26. These results imply that GO is a potentially ideal nanomaterial for improving the efficacy of anti-cancer drugs that function through intercalating to nucleic acid. In addition to that, we also found that the lateral dimension of GO sheets is a critical factor for the cleavage activity of GO/Cu2+ system. This is expected because the diameter of normal B-DNA helix is much smaller comparing to the GO sheets with the lateral size range from several nanometer to micrometer. Possibly only those GO sheets with the comparable size to DNA interact with it. We, hence, developed recently a facile preparation method of graphene quantum dots (GQDs) with periphery carboxylic groups and average lateral size of ~30 nm or even smaller. Due to their unique structural property, the as-prepared GQDs dispersed very well in aqueous solution. Combining with copper ions, GQDs exhibited several folds higher DNA cleavage activity than that of GO27. Therefore, we foresee that the GQDs might improve the nuclease activity of the chemical nucleases. Figure 1a compared the DNA cleavage activity enhancements by GQDs (average lateral size, ~20 nm) and GO to a classical chemical nuclease, di-1,10-phenanthroline-copper (Cu(phen)2). It was well known that increasing the aromatic plane of Cu(phen)2 could improve its DNA cleavage activity through increasing its affinity to DNA28. As expected, under the same reaction conditions, the supercoiled plasmid DNA (Band I) can be cleaved more completely in the presence of the GQDs than that of GO (lane 2 vs lane 10). At a high concentration of GQDs, not only nicked DNA (Band II), linear DNA (Band III) were also generated; whereas for GO, only nicked DNA fragments were observed (lanes 7–12 vs lanes 1–6). Additionally, no DNA samples dwelled in the sample wells in the case of GQDs even at a higher concentration indicates that GQDs also have a better size consistence and biocompatibility27.
a) DNA (38 μM bp) cleavage by Cu(phen)2 (di-1,10-phenanthroline-copper) (1 mM) with GO and GQDs. The reactions were performed at 37°C for 1 h. Lanes 1–6: the concentrations of GO were 10, 30, 50, 70, 100 and 150 μg mL−1 Lanes 7–12: GQDs were 2, 5, 10, 30, 50 and 70 μg mL−1. The first two lanes were the controls of DNA alone and DNA with Cu(phen)2 (1 mM). b) Cleavage of the DNA (38 μM bp) with DOX (10 μM), NaBH4 (25 mM) and different amounts of GQDs as labeled. Reaction time was 1 h (37°C). Lanes 1 and 2 are the controls of DNA, DNA and NaBH4 (25 mM).
On the basis of the aforementioned results, we hypothesize that GQDs will show high potency in cancer therapy than GO. The assumption is examined systematically in this work using an anti-cancer drug doxorubicin (DOX). The interaction of GQD with DOX, cellular internalization, cellular distribution and cytotoxicity of the GQDs and potential application in drug-resistant cells were investigated. We found that without any pre-modification, GQDs not only can efficiently accelerate DOX nuclear accumulation, but also enhance markedly the DNA cleavage activity and cytotoxicity of DOX, which are superior to the modified graphene or GO and many nanoparticle-based DOX delivery systems8,9,10,11.
Results
Enhanced DNA cleavage activity of DOX by the GQDs
To discover the potential of the GQDs in anti-cancer therapeutics, the effect of GQDs on the DNA cleavage of DOX was explored initially. DOX is one of the most commonly used anti-cancer chemotherapeutics, but its clinical utility is limited by low solubility and severe side effects29. The cytotoxicity of DOX is associated with its intercalation between two base-pairs of DNA in the nucleus to form DNA adduct or DNA cross-linking inducing interference with DNA strand separation and DNA helicase30. DOX is also believed to be reduced to a semiquinone radical inside cell, which is generated in electron-transfer chains and induces the formation of highly reactive hydroxyl radicals that capable of cleaving DNA molecules31,32. Figure 1b shows that at a low concentration of GQDs, the DNA cleavage by DOX is barely affected; at about 60 μg mL−1 of GQDs, the percentage of nicked DNA fragment starts to increase and cleavage activity is improved and reaches the maximum with 150 μg mL−1 of GQDs, further increasing of the GQDs concentration results in a decline in the cleavage activity. It was also noticed that the DNA cleavage enhancement by GQDs is time dependent. Increasing the incubation time, the cleavage is more complete as shown in Figure S1. In the presence of 150 μg mL−1 of GQDs, after 2 h of incubation, the plasmid DNA can be cleaved completely into nicked (98%) and linear (2%) DNA fragments. The time and concentration dependent cleavage phenomena unambiguously show that the GQDs can enhance DNA double strand breaks. Noticeably, the activity enhancement is critically dependent on the ratio between GQD and DOX. This observation is analogous to the behavior of the GO/chemical nuclease conjugates26, suggesting that GQDs possibly work together with DOX molecules.
Formation of DOX and GQD conjugates (DOX/GQD)
To explore the state of the DOX and GQD in solution, the electronic and fluorescence properties of DOX were employed to monitor their interactions. Figure 2a depicted the changes in the UV-vis spectrum of DOX in the presence of different amount of GQD. Decrease of the peak intensity at 490 nm that ascribed to the π-π transitions of electrons of the aromatic rings of DOX with the increase of the concentration of GQD suggests that a π-π interaction occurred between the GQD and DOX as observed for the interaction between GO and DOX24, because the GQD assumes a planar structure with peripheral carboxylic groups that is similar to GO. Apparent binding constant (Kb, 0.013 μg mL−1) of the DOX to GQD obtained using the previously published procedure33 was smaller compared to the binding constants of the chemical nuclases to GO, possibly because the electrostatic interaction contributed less in the case of GQDs27. Nevertheless, the results suggested that GQDs and DOX molecules formed conjugates in solution. To verify the conjugation state of the DOX with GQD, the fluorescence of DOX was monitored in the presence of different amount of GQDs. It was reported that the fluorescence of small molecules can be quenched by the GO because of their π-π stacking with GO23,34. Hence the change of the fluorescence of DOX can be used to monitor its interaction with GQDs. As expected, the fluorescence intensity of DOX decreased with the increase of the concentration of GQDs and was mostly quenched at DOX to GQD ratio of ~0.04 (μg to μg) (Figure 2b). Most noticeably, this ratio was consistent with the ratio, at which the DNA cleavage activity of DOX was enhanced maximally (Figure 1b). The ratio thus was used hereafter to prepare DOX/GQD conjugates assuming that there is no free DOX under this condition. The conjugation state was also confirmed by their AFM images as shown in Figure S2. The thickness of the GQDs is obviously increased from ~1 nm to ~3 nm when forming DOX/GQD conjugates. The formation of the conjugates was also supported by the changes in the zeta potential of the GQDs in the presence of DOX (Figure S3). The stability of the DOX/GQD conjugate was tested for different storage time and in the buffers with different pH values and ionic strengths. The conjugates are stable at room temperature for 36 h in PBS buffer or cellular culture medium (Figure S4a, b), no changes occurred in the solutions with different ionic strengths neither (Figure S4c), just a slight fluorescence variation in the solutions from pH 2 to 7.4 was observed (Figure S4d). The later is different from that of the DOX/GO system, in which ~70% DOX could be released at pH 2 after 24 h35. On the basis of these results, a molecular model of the DOX/GQD conjugate was built as shown in Figure S5. The high stability of the DOX/GQD conjugates further supports that the interaction between GQD and DOX is mainly contributed by π-π stacking. This conclusion was solidified complementarily by the complete release of DOX from GQDs in 50% ethanol as shown in Figure S5.
a) Absorption spectra of the DOX (40 μM) with increasing GQDs concentrations (from 5 to 75 μg mL−1). Inset is the plot of [GQDs]/(εa − εf) versus GQDs concentration, straight line is the linear fit of the plot. εa and εf were described in the Experimental section. b) Fluorescence of the DOX (20 μM) was quenched by the increasing amount of GQDs (from 20 to 300 μg mL−1) in 10 mM PBS buffer (pH 7.4). Excitation wavelength was 500 nm. c) Cell viability of MCF-7 cells after 24 h exposure to 10 μM of DOX in the presence of different concentrations of GQDs at 37°C. Zero spot indicates without GQDs, corresponding to the point that circled in the inset. Inset is the cell viability of the MCF-7 cells after 24 h incubation with different concentrations of DOX alone at 37°C.
Enhance cytotoxicity of DOX by GQDs
The high stability of the DOX/GQD conjugates implies that the function of GQDs in the interaction of DOX with DNA molecules is similar to GO/chemical nuclease systems27. The GQDs increased the binding affinity of DOX to DNA, eventually improved its DNA cleavage activity. It is well known that DNA cleavage by DOX and its derivatives normally results in an enhanced cytotoxic potency36. In attempt to understand whether the enhancement in DNA cleavage activity of DOX by GQDs is correlated to cytotoxicity, the impact of DOX/GQD conjugates on the cytotoxicity of DOX to human breast cancer cells MCF-7 and kidney cancer cells MGC-803 was studied. DOX alone exhibits cytotoxicity to both MCF-7 and MGC-803 cells, insets in Figures 2c and S6, as reported in the literature37,38. The GQDs alone under the same conditions showed very low toxicity to both cell lines (Figure S7)39. In the presence of GQDs, cytotoxicity of DOX to both cell lines was enhanced,which was increased with the increase of the concentration of GQDs (Figures 2c and S6). In the case of MCF-7, the cell viability was dropped from 58% to 35% with the presence of 50 μg mL−1 of GQDs. The viability was then slightly lowered with the further increase of GQDs concentration. Similar to MCF-7 cells, the cell viability of MGC-803 cells lines also decreased noticeably starting from 50 μg mL−1 of GQDs. Further increase of the GQDs, the cytotoxicity maintains unchanged. The improved cell killing in the presence of GQDs is consistent with the observation for the DOX derivatives, in which high DNA cleavage activity corresponds to an increased cytotoxic potency36. Though the correlation of the DNA cleavage activity with the cytotoxicity was actually reported for the anti-cancer drugs encapsulated in polymer-lipid hybrid nanoparticle40, it is necessary to clarify the intracellular distribution of the DOX/GQD conjugates first to solidify the result.
Effects of GQDs on the cellular uptake and intracellular distribution of DOX
Although the DOX/GQD conjugates are stable in cell culture media (Figure S4), the status of the DOX/GQD conjugates inside the cell is still a key issue that needs to be illustrated in order to correlate their nuclease activity and cytotoxicity. To understand this question, confocal laser scanning microscopy (CLSM) images of the cells incubated with DOX, GQDs and DOX/GQDs for different incubation times were taken, respectively, as shown in Figure 3. The impact of the DOX/GQD conjugates is compared with the free DOX and GQDs in terms of the fluorescence intensity and distribution. Fluorescence intensity of the as-prepared GQDs is relative weak27 and also the fluorescence emission wavelength of GQDs overlaps with the cellular auto fluorescence, thus the red fluorescence of DOX was monitored instead. With the incubation time increasing from 4 to 24 h (comparing vertically in Figure 3), the fluorescence intensity of the cells with GQDs alone was barely increased. The fluorescence of the cells with DOX alone increases with the incubation time and DOX molecules gradually diffuse into nuclei after 24 h of incubation, consistent with the results in the literature10,12. Noticeably, the fluorescence of the cells incubated with DOX/GQD conjugates for 4 h originates exclusively from the nuclei. Almost no fluorescence was observed in the cytoplasm, indicating that free DOX molecules solely located inside the nucleus under this condition. In addition, the fluorescence intensity of the cells that incubated with DOX/GQD is about two folds higher than that of the cells incubated with DOX alone under the same concentration (right column in Figure 3 and Figure S8). The rapid nuclear uptake and stronger fluorescence intensity in the nuclei reveal undoubtedly that DOX/GQD conjugates increase the rate and amount of the DOX nuclear accumulation. To further confirm the result, the nuclei were stained with Hoechst and the images were overlapped with the DOX fluorescence images as shown in Figure S9. The overlays (panel d in Figure S9) of the stained nuclei, DOX and the bright field images clearly corroborate the result that DOX accumulation occurs in the nucleus solely in the presence of GQDs. Remarkably, the enhancement on the DOX penetration into nuclei is also dependent on the concentration of GQDs, higher concentration of GQDs is favorable to the DOX nuclear uptake (panel c in Figure S9, comparing horizontally).
CLSM images of the MCF-7 cells incubated with GQDs (15 μg mL−1), DOX (1 μM) and DOX/GQD conjugates under the same corresponding DOX and GQD concentrations for 4, 12 and 24 h excited at 488 nm.
Scale bar: 50 μm. The fourth row shows the DOX nuclear accumulation of the MCF-7 cells incubated with DOX (1 μM) and DOX (1 μM) with GQDs (15 μg mL−1) under the same concentration for 4, 12 and 24 h. The fluorescence intensities were generated by Image-Pro Plus program.
The nuclear accumulation enhancement by GQDs was further proved by flow cytometry, in which the cellular uptake of DOX under different conditions can be compared quantitatively. Figure 4 displayed the cellular uptakes of the DOX alone and DOX/GQD conjugates incubated for 1 and 4 h, respectively. With a short incubation time (1 h, top panel in Figure 4), cytometric profiles clearly indicated that fluorescence intensity of the cells with the DOX/GQD conjugates is stronger (red peak) than that with DOX alone (blue peak), which is consistent with the corresponding CLSM images on the right. With the incubation time increase up to 4 h, more DOX molecules were accumulated inside the cell in both cases and the difference in the total fluorescence intensity between the two samples is getting smaller with a longer incubation time. However, for the cells incubated with DOX, the fluorescence intensity is generated by the DOX distributed both in the cytoplasm and nucleus of the cells. While for the cells incubated with the DOX/GQD conjugates, the total fluorescence intensity is from the DOX molecules that are inside nuclei only. Hence, even the fluorescence intensities of the cells incubated with DOX alone or DOX/GQD conjugates are comparable after the longer incubation time, the nuclear accumulation of DOX is still higher with DOX/GQD conjugates than DOX alone. The results collectively showed that under the same DOX concentration, the GQDs dramatically enhanced DOX nuclear uptake. This finding is therapeutically more important than the enhanced intracellular uptake. In fact, in many anti-cancer nanodrug systems, cellular uptake of the drugs was usually improved, but few of them could enhance the drug nuclear uptake without nuclear-targeted modification8,9,10,11. In addition, the results may be practically important to reduce the side effects imposed by the high dose of anti-cancer drugs.
Left: Flow cytometric profiles of the MCF-7 cells incubated with DOX (1 μM) alone and DOX/GQDs (1 μM/15 μg mL−1) for 1 and 4 h. The black curves in the flow cytometric profiles are control cells, blue curves are fluorescence of the cells incubated with DOX alone and red curves are fluorescence of the cells with the DOX/GQDs. Right: The fluorescence images of the corresponding samples. Scale bar: 50 μm.
Cellular internalization and functional mechanism of the DOX/GQD conjugates
Generally, cells uptake exogenous materials such as nanoparticles through endocytosis41,42. We previously found that both energy-independent and energy-dependent pathways are possibly involved in the GQDs cellular uptake and caveolae-mediated endocytosis may be the primary pathway39. Figure S10 showed the fluorescence images of the cells that were pretreated with different inhibitors of various endocytotic mechanisms including chlorpromazine, filipin III and NaN3 and then incubated with the DOX/GQD conjugates for 4 h. The fluorescence intensities of the cells pretreated with three inhibitors are all weakened with different extents and among them chlorpromazine seems relatively more effective as shown in panels b and d in Figure S10. The results indicate that energy-dependent endocytosis, clathrin-mediated endocytosis and caveolae-mediated endocytosis are all likely involved in the cellular uptake of the DOX/GQD conjugates. Multiple endocytotic pathways may contribute partially to the efficient cellular uptake of DOX via DOX/GQD conjugates.
Through endocytosis, cells eventually route some exogenic materials towards lysosomes where the enzymes will degrade them to protect cells. As showed in Figure S4, the DOX/GQD conjugates is very stable from pH 2–10, suggesting that pH trigged DOX release is unlikely to occur in acidic lysosome43,44. The release of DOX from the DOX/GQDs conjugates may take place in a hydrophobic environment, because the fluorescence of DOX that quenched by GQDs can be completely recovered when adding 50% ethanol (Figure S5). However, such strong hydrophobic environment is generally unlikely existed inside cells. It was well known that the particles that are smaller than ~10 nm can freely diffuse into the nucleus45. Owing to the inherent flexibility and small size of the GQDs, it is very likely DOX/GQD conjugates penetrate the nucleus by diffusion. DNA-polylysine complexes as large as ~60 nm were shown to efficiently deliver DNA to the nucleus, in which it was believed that the larger structures were able to pass through the nuclear pores owing to the inherent flexibility of the conjugate46. Hence, we postulate that the release of DOX from the DOX/GQD conjugates occur inside the nucleus when they interact with DNA molecules. This hypothesis is consistent with the observation in the fluorescence microscopy images. No fluorescence of DOX was observed in the cytoplasm in the presence of GQDs (Figures 3, 4 and S9) indicates that DOX molecules exist as DOX/GQD conjugates in the cytoplasm, because the fluorescence of DOX will be quenched by GQDs when they bind together (Figure 2b). The strong fluorescence intensity of DOX displayed in the nuclei when the cells incubated with DOX/GQDs even for a shorter incubation time (1 h in Figure 4), confirmed that DOX separated from GQDs in the nucleus. The release of DOX from the DOX/GQD conjugates in the nucleus was also supported by the DOX efflux experiment. Figure S11 showed the fluorescence microscopy images of the cells incubated with the DOX/GQDs first, then continuously incubated with fresh media without GQD or DOX. With a longer incubation time, DOX molecules start to diffuse to the cytoplasm, indicating the DOX molecules that entered as the DOX/GQD conjugates are free in the nuclei. We also found that once DOX binds to DNA, they would not bind to the GQD anymore. (Figure S12). The detachment of DOX from the DOX/GQD conjugates in nucleus was also strongly supported by the fluorescence diminish of the pre-stained nuclei after the incubation with GQDs as shown in Figure 5. With the concentration of GQDs increase, the fluorescence of nuclei gradually decreased. This could be caused either by the disruption of the interaction between the dye and DNA, or fluorescence was quenched by GQDs directly. Nevertheless, the result revealed unanimously that GQDs entered into the nuclei. Taken all these results together, the cellular and nuclear uptake mechanism of the DOX/GQDs was schematically summarized in Figure 6. The DOX/GQD conjugates entered the cell through clathrin-mediated and caveolae-mediated endocytosis, then were routed to lysosomes or Golgi before diffusing into nucleus39. The release of DOX from the conjugates occurs inside nucleus driven by the higher affinity of DOX/GQDs towards DNA molecules, though it is hard to detect the DOX binding to DNA in cell nuclei47. Our data, although not sufficient to directly support every step of this process, are consistent with such an interpretation. The release mechanism of the DOX from DOX/GQD conjugate in vivo requires further investigation.
Discussion
The superb performance of the DOX/GQD conjugates in targeted drug delivery, DNA cleavage and cytotoxicity to MCF-7 cells suggests that the conjugates may be capable of suppressing the cellular resistance to DOX. As one of the generally accepted drug-resistant mechanisms is that the membrane bound active drug efflux pumps are over expressed in many drug-resistant cells48,49. Bypassing these transporters via DOX/GQD conjugates can therefore possibly avoid the DOX resistance. To test this hypothesis, the cellular penetration of GQDs alone to the DOX resistant cells MCF-7/ADR that were exposed to a low dose of DOX (1 μM), was first tested (Figure S13). The GQDs can easily enter the MCF-7/ADR cells as indicated by the quenched fluorescence of DOX in the cytoplasm. However, the entered DOX molecules only dwell in the cytoplasm, even with a higher DOX concentration (Figure 7, DOX column) that agreed with the literature data (Figure 7, control column)50,51. When the MCF-7/ADR cells were incubated with the DOX/GQD conjugates, remarkably, instead of residing in the cytoplasm, DOX penetrates into the nuclei (Figure 7, Top part, DOX/GQD column). The results are more obvious when the images are overlapped with the bright field images, the Hoechest dyed nuclear image and DOX fluorescence as shown in the panel d of Figure 7. This nuclear penetration is in accordance with the cytotoxicity enhancement of the DOX/GQDs to MCF-7/ADR cells as shown in the bottom part of Figure 7. Under the constant GQD concentration, the cytotoxicity of DOX/GQDs to MCF-7/ADR cells is higher than that of free DOX (red line versus black line) indicating DOX was efficiently delivered to the drug resistant cells by DOX/GQD conjugates. We also observed that at a low DOX concentration, the growth of the MCF-7/ADR cells was enhanced by DOX alone, but not by DOX/GQD conjugates. Overall, this preliminary data of the drug resistant cells validates our results performed on the wild type cancer cells. More importantly, the result suggests it is possible to overcome drug resistance in tumor cells using DOX/GQD conjugates.
Top: Fluorescence images of the MCF-7/ADR cells after the incubation n with GQDs, DOX and DOX/GQDs. a) bright-field, b) nuclei stained by Hoechst, c) fluorescence of DOX excited at 510 nm and d) overlays of a, b and c. Scale bar: 50 μm. Bottom: Cell viability of the MCF-7/ADR cells exposed to different concentrations of DOX alone (dark bars) and DOX with GQDs (100 μg mL−1) for 24 h (grey bars). The control samples are the untreated cells and the cells with GQDs only (100 μg mL−1, gray bar).
In summary, we demonstrate that the GQDs possess dual function as a targeted anti-cancer drug carrier and DNA cleavage activity enhancer, which should be potentially useful in cancer therapy. The GQDs interact with DOX forming stable conjugates, which deliver their cargo to the nucleus specifically and simultaneously improve the DNA cleavage activity of DOX. The enhancement of the GQDs on the DNA cleavage activity and nuclear accumulation of DOX lead to its higher cytotoxicity both to wild type and drug-resistant cancer cells. The results are encouraging, this is the type of nanomaterials that could accelerate efficiently the nuclear accumulation of the anti-cancer drug and simultaneously enhance the anti-tumor efficacy of the drug without any pre-modification with chemicals or biological molecules. The results are practically important, the GQD conjugates might be used to increase chemotherapy efficacy, to reduce the side effects imposed by the high dose of anti-cancer drugs and more significantly to improve suboptimal therapeutic response of the anti-cancer drugs that associated with the drug resistance.
Methods
General information
DOX was purchased from Aladdin Chemistry Co. Ltd. Supercoiled PSICOR-GFP DNA plasmid was purified from DH5α cells using an Endo Free Plasmid Kit (QIAGEN, USA). All other chemicals and reagents were purchased from Sino Pharm Chemical Reagent Co. Ltd. in analytic grade and were used as received.
Agarose gel electrophoresis was carried out with a DYY-6C electrophoresis apparatus (Liuyi Instrumental Co., China). The agarose gels were visualized and digitized with the FR-200A gel image analysis system and analyzed by Smart View software. The UV-vis measurements were performed on a Cary 50 spectrometer (Varian, USA). The fluorescence spectra were acquired by a Cary Eclipse spectrofluorometer (Varian, USA). Fluorescence images of cells were obtained by a fluorescence microscopy (Nikon ECLIPSE, Ti-S) and A1R confocal laser scanning microscope (Nikon, Japan). Flow cytometry was performed on FACS Calibur (BD Biosciences, Mountain View, CA).
GQDs preparation and characterization
The GQDs were prepared through photo-Fenton reactions of graphene oxide as described in our previous work27. The photo-Fenton reactions were carried out in a quartz tube with graphene oxide and Fenton reagent, FeCl3 and H2O2 mixture under vigorous stirring in a photo reactor and initiated by a mercury lamp (365 nm, 1000 W) (Bilon, Shanghai). The pH of the mixture was adjusted to 4. The reaction products were dialyzed in ultrapure water for two days to remove reactants and small product molecules. The generated GQDs were characterized by AFM, XPS, TEM and FT-IR, UV-visible and Florescence spectroscopies. The aqueous suspension of GQD was stored at room temperature and sonicated, filtered with a sterilized 0.22 μm filter prior to use.
DNA cleavage experiments
DNA cleavage activity of DOX was studied by agarose gel electrophoresis. Excess and freshly prepared NaBH4 solution was used as reducing agent32. Reactions were carried out in sodium phosphate buffer (50 mM, pH 7.0). The DNA plasmid was incubated with different concentration of DOX and GQD at 37°C, quenched by addition of the loading buffer (0.05% bromophenol blue, 1% SDS and 50% glycerol). The mixture was then subjected to electrophoresis on a 0.5% agarose gel containing ethidum bromide (0.5 μg mL−1) in TBE buffer at 90 V for approximately 1 h25,26.
Binding measurements of DOX to GQD
DOX and GQD aqueous solutions were mixed sufficiently and incubated for 5 min in dark; the UV-vis absorption of the mixture was then recorded. The binding constant of DOX to GQD was calculated according to the previously published method33. Fluorescence spectroscopy was also employed to investigate the interaction of DOX with GQD. Fluorescence spectrum of DOX were recorded by using an excitation wavelength of 500 nm with the increased concentrations of GQD.
Cell culture
Human breast cancer cells MCF-7 and gastric cancer MGC-803 cells (purchased from Shanghai Cell Bank of the Chinese Academy of Sciences) were cultured in RPMI 1640 medium with 10% fetal bovine serum and 1% antibiotics (penicillin and streptomycin) at 37°C under 5% CO2. DOX resistant cells MCF-7/ADR (purchased from Ruizhi Bio-pharmaceutical Technology Co., Ltd) were cultured under the same condition, except for with 1 μM of DOX to maintain their drug resistance.
Fluorescence microscopy and confocal laser scanning microscopy
MCF-7 cells were detached by trypsin and plated on the collagen coated Φ14 mm cover slips at a density of 5 × 104 cells per well in 24-well plates to culture sequentially. After 12 h of attaching, the medium was replaced with the medium without serum but containing 1 μM of DOX or 1 μM of DOX with 150 μg mL−1 of GQD followed by the washing with PBS buffer two times. For comparison the control cells were incubated in the fresh medium without serum. After incubation for different time, the cells were washed two times with PBS buffer to remove free drug and GQDs and then fixed with 4% paraformaldehyde (pH = 7.4). After 15 min for fixing, the paraformaldehyde was removed10. The cells were washed two times with PBS buffer to remove the residual fixing reagent. To stain the nuclei, 300 μL of Hoechst PBS solution (0.5 μg mL−1) was added at this stage and incubated for 5 min7. The coverslips were laid onto slides after washing with PBS buffer. The prepared slides were imaged by fluorescence microscope or confocal laser scanning microscope.
Cytotoxicity assay
The viability of MCF-7 and MGC-803 cells in the presence of DOX or DOX/GQDs was assayed using MTT assay kit (Beyotime Institute of Biotechnology, China). Cells were plated in a 96 well microplate at a density of 4000–5000 cells. Background control wells containing the same volume of complete culture medium were included in each assay. The microplate was incubated for 24 h at 37°C. Then the cells were washed with PBS buffer and incubated with DOX and GQDs in serum-free medium and the plate was incubated further for 24 h. The optical density of DMSO solution of formazan at 490 nm was recorded after 4 h of incubation with MTT12.
Flow cytometry measurement
To obtain a quantitative cellular uptake of DOX in the presence of GQDs, MCF-7 cells were cultured with DOX or DOX/GQD in a 6-well plate at the density of 5 × 105 cells per well for different time. The cells were then washed with PBS three times and harvested. The cells were re-suspended in PBS buffer (10 mM) and fixed with cold 70% ethanol aqueous solution overnight for flow cytometric measurement51.
References
Devadasu, V. R., Bhardwaj, V. & Kumar, M. N. V. R. Can Controversial Nanotechnology Promise Drug Delivery? Chem. Rev. 113, 1686–1735 (2012).
Peer, D. et al. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotech. 2, 751–760 (2007).
Prato, M., Kostarelos, K. & Bianco, A. Functionalized Carbon Nanotubes in Drug Design and Discovery. Acc. Chem. Res. 41, 60–68 (2007).
Mane, S. R. et al. Amphiphilic Homopolymer Vesicles as Unique Nano-Carriers for Cancer Therapy. Macromolecules 45, 8037–8042 (2012).
Zhou, H. et al. Synthesis and Characterization of Amphiphilic Glycidol−Chitosan−Deoxycholic Acid Nanoparticles as a Drug Carrier for Doxorubicin. Biomacromolecules 11, 3480–3486 (2010).
Tkachenko, A. G. et al. Multifunctional Gold Nanoparticle−Peptide Complexes for Nuclear Targeting. J. Am. Chem. Soc. 125, 4700–4701 (2003).
Song, J., Zhou, J. & Duan, H. Self-Assembled Plasmonic Vesicles of SERS-Encoded Amphiphilic Gold Nanoparticles for Cancer Cell Targeting and Traceable Intracellular Drug Delivery. J. Am. Chem. Soc. 134, 13458–13469 (2012).
Kim, J. et al. Designed Fabrication of a Multifunctional Polymer Nanomedical Platform for Simultaneous Cancer-Targeted Imaging and Magnetically Guided Drug Delivery. Adv. Mater. 20, 478–483 (2008).
Muhammad, F. et al. pH-Triggered Controlled Drug Release from Mesoporous Silica Nanoparticles via Intracelluar Dissolution of ZnO Nanolids. J. Am. Chem. Soc. 133, 8778–8781 (2011).
Zhao, Y. X. et al. DNA Origami Delivery System for Cancer Therapy with Tunable Release Properties. ACS Nano 6, 8684–8691 (2012).
Paliwal, S. R. et al. Estrogen-Anchored pH-Sensitive Liposomes as Nanomodule Designed for Site-Specific Delivery of Doxorubicin in Breast Cancer Therapy. Mol. Pharm. 9, 176–186 (2011).
Pan, L. et al. Nuclear-Targeted Drug Delivery of TAT Peptide-Conjugated Monodisperse Mesoporous Silica Nanoparticles. J. Am. Chem. Soc. 134, 5722–5725 (2012).
K, C., R, B., Thapa, B. & Xu, P. pH and Redox Dual Responsive Nanoparticle for Nuclear Targeted Drug Delivery. Mol. Pharm. 9, 2719–2729 (2012).
Dobrovolskaia, M. A. & McNeil, S. E. Immunological properties of engineered nanomaterials. Nature Nanotech. 2, 469–478 (2007).
Mao, H. Y. et al. Graphene: Promises, Facts, Opportunities and Challenges in Nanomedicine. Chem. Rev. 113, 3407–3424 (2013).
Feng, L. & Liu, Z. Graphene in biomedicine: opportunities and challenges. Nanomedicine 6, 317–324 (2011).
Zhang, Y., Nayak, T. R., Hong, H. & Cai, W. Graphene: a versatile nanoplatform for biomedical applications. Nanoscale 4, 3833–3842 (2012).
Sun, X. et al. Nano-graphene oxide for cellular imaging and drug delivery. Nano Res. 1, 203–212 (2008).
Zhou, L. et al. Graphene Oxide Noncovalent Photosensitizer and Its Anticancer Activity In Vitro. Chem. Eur. J. 17, 12084–12091 (2011).
Huang, P. et al. Folic Acid-conjugated Graphene Oxide loaded with Photosensitizers for Targeting Photodynamic Therapy. Theranostics 1, 240–250 (2011).
Yang, X. et al. Multi-functionalized graphene oxide based anticancer drug-carrier with dual-targeting function and pH-sensitivity. J. Mater. Chem. 21, 3448–3454 (2011).
Yang, K. et al. Graphene in Mice: Ultrahigh In Vivo Tumor Uptake and Efficient Photothermal Therapy. Nano Lett. 10, 3318–3323 (2010).
Liu, Z., Robinson, J. T., Sun, X. & Dai, H. PEGylated Nanographene Oxide for Delivery of Water-Insoluble Cancer Drugs. J. Am. Chem. Soc. 130, 10876–10877 (2008).
Zhang, L., Xia, J., Zhao, Q., Liu, L. & Zhang, Z. Functional Graphene Oxide as a Nanocarrier for Controlled Loading and Targeted Delivery of Mixed Anticancer Drugs. Small 6, 537–544 (2010).
Ren, H. et al. DNA Cleavage System of Nanosized Graphene Oxide Sheets and Copper Ions. ACS Nano 4, 7169–7174 (2010).
Zheng, B. et al. Nuclease Activity and Cytotoxicity Enhancement of the DNA Intercalators via Graphene Oxide. J. Phys. Chem. C 116, 15839–15846 (2012).
Zhou, X. et al. Photo-Fenton Reaction of Graphene Oxide: A New Strategy to Prepare Graphene Quantum Dots for DNA Cleavage. ACS Nano 6, 6592–6599 (2012).
David, S., Sigman, R. L., David, M. & Perrin, L. P. Nucleic Acid Chemistry of the Cuprous Complexes of 1,10-Phenanthroline and Derivatives (Institute of Inorganic Chemistry, University of Basel, Place, 1996).
Carter, S. K. Adriamycin–A Review. J. Natl. Cancer Inst. 55, 1263–1263 (1975).
Swift, L. P., Rephaeli, A., Nudelman, A., Phillips, D. R. & Cutts, S. M. Doxorubicin-DNA Adducts Induce a Non-Topoisomerase II–Mediated Form of Cell Death. Cancer Res. 66, 4863–4871 (2006).
Feinstein, E., Canaani, E. & Weiner, L. M. Dependence of Nucleic Acid Degradation on in situ Free-Radical Production by Adriamycin. Biochemistry 32, 13156–13161 (1993).
William Lown, J., Sim, S. K., Majumdar, K. C. & Chang, R. Y. Strand scission of DNA by bound adriamycin and daunorubicin in the presence of reducing agents. Biochem. Biophys. Res. Commun. 76, 705–710 (1977).
Kalsbeck, W. A. & Thorp, H. H. Determining Binding Constants of Metal Complexes to DNA by Quenching of the Emission of Pt2(pop)44- (pop = P2O5H22-). J. Am. Chem. Soc. 115, 7146–7151 (1993).
Liu, Z., Sun, X., Nakayama-Ratchford, N. & Dai, H. Supramolecular Chemistry on Water-Soluble Carbon Nanotubes for Drug Loading and Delivery. ACS Nano 1, 50–56 (2007).
Yang, X. et al. High-Efficiency Loading and Controlled Release of Doxorubicin Hydrochloride on Graphene Oxide. J. Phys. Chem. C 112, 17554–17558 (2008).
Capranico, G., Soranzo, C. & Zunino, F. Single-Strand DNA Breaks Induced by Chromophore-modified Anthracyclines in P388 Leukemia Cells. Cancer Res. 46, 5499–5503 (1986).
Ravid, A. et al. 1,25-Dihydroxyvitamin D3 Enhances the Susceptibility of Breast Cancer Cells to Doxorubicin-induced Oxidative Damage. Cancer Res. 59, 862–867 (1999).
Xi, J., Zhou, L. & Dai, H. Drug-loaded chondroitin sulfate-based nanogels: Preparation and characterization. Colloids Surf. B Biointerfaces 100, 107–115 (2012).
Wu, C. et al. Cellular Internalization and Low Cytotoxicity of Graphene Quantum Dots. Adv. Healthcare Mater. 10.1002/adhm.201300066 (2013).
Shuhendler, A. et al. A novel doxorubicin-mitomycin C co-encapsulated nanoparticle formulation exhibits anti-cancer synergy in multidrug resistant human breast cancer cells. Breast Cancer Res. Treat. 119, 255–269 (2010).
Zaki, N. M. & Tirelli, N. Gateways for the intracellular access of nanocarriers: a review of receptor-mediated endocytosis mechanisms and of strategies in receptor targeting. Expert Opin. Drug Deliv. 7, 895–913 (2010).
Mu, Q. et al. Size-Dependent Cell Uptake of Protein-Coated Graphene Oxide Nanosheets. ACS Appl. Mater. Interfaces 4, 2259–2266 (2012).
Du, J. Z., Du, X. J., Mao, C. Q. & Wang, J. Tailor-Made Dual pH-Sensitive Polymer–Doxorubicin Nanoparticles for Efficient Anticancer Drug Delivery. J. Am. Chem. Soc. 133, 17560–17563 (2011).
Su, J., Chen, F., Cryns, V. L. & Messersmith, P. B. Catechol Polymers for pH-Responsive, Targeted Drug Delivery to Cancer Cells. J. Am. Chem. Soc. 133, 11850–11853 (2011).
Paine, P. L., Moore, L. C. & Horowitz, S. B. Nuclear envelope permeability. Nature 254, 109–114 (1975).
Chan, C. K. & Jans, D. A. Supramolecular structure and nuclear targeting efficiency determine the enhancement of transfection by modifed polylysines. Gene Ther. 7, 1690–1697 (2000).
Lam, W., Leung, C. H., Chan, H. L. & Fong, W. F. Toxicity and DNA binding of dextran-doxorubicin conjugates in multidrug-resistant KB-V1 cells: optimization of dextran size. Anti-Cancer Drug 11, 377–384 (2000).
Gottesman, M. M., Fojo, T. & Bates, S. E. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat. Rev. Cancer 2, 48–58 (2002).
Gottesman, M. M. Mechanism Of Cancer Drug Resistance. Annu. Rev. Med. 53, 615–627 (2002).
Jiang, Q. et al. DNA Origami as a Carrier for Circumvention of Drug Resistance. J. Am. Chem. Soc. 134, 13396–13403 (2012).
Chen, Y., Bathula, S. R., Li, J. & Huang, L. Multifunctional Nanoparticles Delivering Small Interfering RNA and Doxorubicin Overcome Drug Resistance in Cancer. J. Biol. Chem. 285, 22639–22650 (2010).
Acknowledgements
This research was carried out with financial support from the National Science foundation of China (Nos. 91123011, 90923041 and 31070742), the state key laboratory of bioreactor engineering (No. 2060204), 111 Project (No. B07023), the Shanghai Committee of Science and Technology (Nos. 11DZ2260600 and 12nm0503500) and National “973 program” (No. 20101CB933900) and “863 program” (No. 2012AA022603) of China.
Author information
Authors and Affiliations
Contributions
J. Zhang and S. Guo designed the project. C. Wang carried out the molecular level experiments. C. Wang and C. Wu performed cellular level experiments. X. Zhou, T. Han, X. Xin and J. Wu prepared and characterized the materials. J. Zhang, S. Guo and C. Wang wrote the manuscript. All authors discussed the results and commented on the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
supplementary dataset
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Wang, C., Wu, C., Zhou, X. et al. Enhancing Cell Nucleus Accumulation and DNA Cleavage Activity of Anti-Cancer Drug via Graphene Quantum Dots. Sci Rep 3, 2852 (2013). https://doi.org/10.1038/srep02852
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02852
This article is cited by
-
Antimicrobial activities of graphene oxide against biofilm and intracellular Staphylococcus aureus isolated from bovine mastitis
BMC Veterinary Research (2023)
-
Biocompatible nucleus-targeted graphene quantum dots for selective killing of cancer cells via DNA damage
Communications Biology (2021)
-
Graphene-based nanomaterial system: a boon in the era of smart nanocarriers
Journal of Pharmaceutical Investigation (2021)
-
Intracellular biosynthesis of PbS quantum dots using Pseudomonas aeruginosa ATCC 27853: evaluation of antibacterial effects and DNA cleavage activities
World Journal of Microbiology and Biotechnology (2020)
-
Recent advances in quantum dots for biomedical applications
Journal of Pharmaceutical Investigation (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.