Abstract
Although RAD52 plays a critical role in the initiation of homologous recombination (HR) by facilitating the replacement of RPA with RAD51, the mechanism controlling RAD52 remains elusive. Here, we show that Bag101, a factor implicated in proteasome functioning, regulates RAD52 protein levels and subsequent HR. LC-MS/MS analysis identified Bag101 which binds to Rad22, the fission yeast homologue of RAD52. Bag101 reduced HR frequency through its overexpression and conversely, HR frequencies were enhanced when it was deleted. Consistent with this observation, Rad22 protein levels was reduced in cells where bag101 was overexpressed even when Rad22 transcription was up-regulated, suggesting the operation of proteasome-mediated Rad22 degradation. Indeed, Rad22 protein levels were stabilized in proteasome mutants. Rad22 physically interacted with the BAG domain of Bag101 and a lack of this domain enhanced HR frequency. Similarly, radiation exposure triggered the dissociation of these proteins so that Rad22 was stabilized and able to enhance HR.
Similar content being viewed by others
Introduction
The stability of the genome is under constant assault from intrinsic and extrinsic stresses, which can lead to DNA damage. Since there are many types of DNA damage, cells possess a number of DNA repair pathways designed to cope with these problems. DNA double-strand breaks (DSBs) which are induced by ionizing radiation or by a stalled replication folk are the most serious type of damage and pose a major threat to cell viability. The presence of DSBs, if even a single DSB remains unrepaired, could lead to mutations and/or chromosomal aberrations and subsequently to cell death or malignancy1. The importance of DSB repair pathways for the maintenance of genomic integrity is underscored by the conservation of these pathways throughout prokaryotes and eukaryotes2. Two major DSB repair pathways are known: non-homologous end joining (NHEJ) and homologous recombination (HR) mediated repair3. In yeast, HR repair is the dominant pathway for DSB repair4,5.
HR repair uses an intact sister chromatid as a substrate, so that it preferentially acts in S phase and G2 phase. HR repair is called an error-free repair pathway for the repair of DSBs. In the absence of HR, DSB repair occurs through NHEJ, even for cells in S phase or G2 phase and this is a more error-prone repair pathway. The undesirable activation of NHEJ leads to an accumulation of mutations and rearrangements in the genome. HR repair is initiated by a DNA resection at the ends of DSBs, providing single-stranded DNA (ssDNA) ends, which are initially coated with RPA, a single-strand binding protein. RPA is subsequently replaced with Rad51, a key recombination enzyme, to form presynaptic nucleofilaments. Next, Rad51 filaments catalyze strand invasion into homologous duplex DNA and DNA polymerases then synthesize DNA over the lesions using a sister chromatid as the template. Finally, ligases rejoin the newly synthesized DNA with the other end of the resected DNA to complete HR repair.
Rad52 is known to promote the replacement of RPA with Rad516. Rad52 forms a homo-heptamer with a ring structure7. The assembly of its ring architecture requires sequences in the evolutionarily conserved N-terminal domain of Rad52, which also possesses DNA binding activity with a preference for ssDNA8. Yeast Rad52 can interact with Rad51 as well as with RPA and facilitates the efficient displacement of RPA on ssDNA by Rad519,10,11. Previous reports indicated that Rad52-deficient yeast cells had severely impaired HR activity, so Rad52 was thought to be a critical regulator of HR repair6.
The HR pathway is strictly controlled, so the disruption of HR activity can induce genomic instability leading to cell death, which is observed in rad51 mutants. These mutants are characterized by a high sensitivity to X-rays and defects in spore formation during meiosis12. Similarly, inappropriate hyper-recombination is associated with genomic instability. For example, the hyper-recombination mutant sgs1 displays higher mitotic and meiotic recombination rates, leading to a short life span for S. cerevisiae13,14. Formation of Rad51 nucleoprotein filaments is a critical step during HR progression and therefore, it is a regulatory factor for HR repair. The UvrD domain helicase, Srs2, prevents HR by disrupting Rad51 nucleoprotein filament formation during S phase15,16,17. During this process, Srs2 is recruited to the site of HR when PCNA is SUMOylated. Similarly, it has been reported that Rad52 is directly SUMOylated on DNA damage sites and regulates HR: it increases intrachromosomal recombination but decreases Rad51-independent recombination18,19,20. During this process, Rad52 is particularly prone to proteasomal degradation and Rad52 SUMOylation increases HR activity by preventing this Rad52 degradation18. These observations suggest that Rad52 degradation contributes to the suppression of HR, both in the absence and presence of DNA damage, although the mechanism of degradation is not yet known.
To investigate the regulatory mechanism involved in Rad52-dependent HR, a search was made for proteins binding to Rad22 (the fission yeast homologue of Rad52) with LC-MS/MS. As a result, sixteen Rad22-binding proteins were detected and their effect on HR activity and Rad22 foci formation was analyzed when they were overexpressed. Among these proteins, attention was focused on Bag101, a fission yeast homologue of the human proteasome associated protein BAG-1. This appeared to be a novel Rad22-binding protein which suppressed HR through Rad22 degradation. Experiments using domain-deleted mutants of Bag101 showed that Bag101 bound to Rad52 through its BAG domain and their dissociation increased HR activity by preventing Rad52 degradation. This dissociation was also observed in cells after IR exposures, demonstrating a novel HR regulatory mechanism operating through Bag101 which is associated with the proteasome.
Results
Rad22-binding proteins regulate HR activity
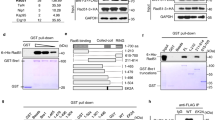
In order to investigate the regulatory mechanisms affecting HR through the actions of Rad22, Rad22-binding proteins were identified with immunoprecipitation using FLATA (FLAG - TEVcs - ProteinA)-tagged Rad22 and LC-MS/MS analysis. Sixteen proteins were co-precipitated from yeast soluble fractions with Rad22-FLATA. These were analyzed with LC-MS/MS using a MASCOT search, which revealed several DNA repair proteins and HSP proteins (Figure 1a). To confirm these interactions in cells, the interactions of Rad22 and the candidate proteins tagged with FH6 (FLAG - 6× His) were examined with a reverse immunoprecipitation method. Rad22-GFP was co-precipitated with each protein, indicating the existence of in vivo interactions (Figure 1b).
Rad22-binding proteins involved in the HR pathway.
(a) Identification of Rad22-binding proteins. Immunoprecipitates were separated on SDS-gradient gels. The bands were visualized with CBB staining. Extracted proteins from each band were identified with LC-MS/MS. (b) Confirmation of in vivo interactions with Rad22. Co-precipitations with anti-FLAG antibodies were performed and immune-blotted with anti-GFP antibody to ensure their interaction between Rad22 and each protein. (c) Spontaneous HR frequency in cells overexpressing each gene. HR was measured by using the RDUX200 reporter as described in Materials and Methods. These experiments were performed three times and error bars represent the S.D. (d) Spontaneous Rad22 focus formation in cells overexpressing each gene. Cells were collected to count the number of GFP-positive cells. Error bars represent the S.D.
Since almost all sixteen proteins interacted with Rad22, it was expected that they would have roles in the regulation of Rad22-associated HR. In order to examine the roles of these proteins in HR, spontaneous HR activity was measured in the clones overexpressing these proteins by using a reporter cassette RDUX200(+). Overexpression of rti1 or rvb2 increased HR frequencies more than 3 times when compared with control cells, while overexpression of bag101, msh2, rim1 or sap1 decreased HR activity (Figure 1c). Since formation of Rad22 foci is associated with Rad22-dependent HR, the formation of Rad22 foci in these overexpressed cells was observed next (See Supplementary Fig. 1). Overexpression of bag101 severely reduced the number of Rad22-foci positive cells when compared to msh2, rim1 or sap1, which is consistent with the reduced frequency of HR (Figures 1c and 1d). On the other hand, overexpression of rvb2 enhanced Rad22-foci positive cells in a pattern, which resembled that of the increased frequency of HR (Figure 1d). Thus Rad22 binding proteins appear to negatively or positively regulate HR activity as observed with Bag101 and Rvb2: Bag101 functions in negative regulation and Rvb2 functions in positive regulation of HR. Bag101 was then examined in an analysis of regulatory mechanisms involved in recombination, because Bag101 is a factor which is associated with the 26S proteasome.
Bag101 deletions increase HR activity and cell viability after IR exposures
To investigate the role of Bag101 in HR after IR exposures, bag101-deletion strains were generated and assayed for IR-induced HR activity. Irradiation with 250 Gy increased HR activity tenfold more than spontaneous levels of HR which were observed in control cells. As expected, overexpression of bag101 reduced IR-induced HR frequencies to half of the level seen in control cells (Figure 2a). Similarly, IR-induced Rad22 focus formation was decreased in bag101-overexpressing cells. Conversely Rad22 focus formation was increased in bag101-deletion cells (Figure 2b). These observations confirmed that bag101 expression negatively regulates Rad22-associated HR.
Association of HR frequency and IR sensitivity with bag101 expression.
(a) IR-induced HR frequency in bag101 overexpressing cells and the deletion mutants. Cells were irradiated with 250 Gy and 500 Gy and the HR frequency was assayed immediately after irradiation. (b) IR-induced Rad22-foci in cells overexpressing Bag101 or containing Bag101 deletions. Cells were irradiated with 250 Gy and 500 Gy and collected 30 minutes later to count Rad22-focus positive cells. (c) IR sensitivity of each cell strain. Cells were irradiated with 250 Gy and 500 Gy and immediately after irradiation, plated on YE plates to form colonies. All experiments in panels (a)–(c) were performed three times and the error bars represent the S.D. (d) The time course of H2A phosphorylation after IR exposure. Cells were irradiated with 500 Gy and at indicated times after irradiation, the lysates from cells overexpressing bag101 (cropped) and deleting bag101 (cropped) were immunoblotted with anti-γ-H2A antibodies under the same condition as that from the wild type cell.
Since HR is the major pathway for repair of IR-induced DSBs, cell viability was measured after IR exposures. Consistent with observations of HR activity, overexpression of bag101 significantly decreased cell viability, while bag101 deletions greatly increased viability (Figure 2c). To determine if these cell viability observations were due to the repair of DSBs, the time course of IR-induced DSB repair was followed with Western blot analysis of the phosphorylation of histone H2A, (i.e. γH2A) which is a marker for the presence of DSBs. γH2A was induced by IR exposures and reached a maximum at 0.2 hour after exposures and then decreased rapidly. In contrast, bag101-overexpressing cells showed a slow decrease in γH2A and retained a significant amount of γH2A at 2 hours post irradiation, although it disappeared in both control and bag101-deleted cells (Figure 2d). These results indicated that Bag101 suppresses HR and the resulting repair of DSBs, leading to decreased cell viability.
Bag101 promotes Rad22 degradation through the proteasome machinery
It has been reported that BAG-1, the human homolog of Bag101, is a coupling factor between Hsc/Hsp70 and the 26S proteasome and accelerates protein degradation26,27. To test this, Rad22 protein levels were measured in each bag101-overexpressing or bag101-deletion cell line. Western blot analyses showed that the amount of Rad22 protein was significantly decreased in bag101-overexpressing cells, while it was robustly increased in bag101-deleted cells (Figure 3a). These observations were consistent with the frequency of Rad22-foci positive cells in non-irradiated and irradiated cells (Figures 1d and 2b). The expression of rad22 mRNA was then measured with quantitative PCR. Unlike the results from the Western Blots, rad22 mRNA levels were not increased in bag101-deletion cells, but were increased in bag101-overexpressing cells (Figure 3b). These results suggest that the Rad22 levels, which were affected by Bag101, were not due to the alteration of Rad22 gene transcription after overexpression or deletion of bag101. Rad22 might have been degraded through the 26S proteasome machinery, because Bag101 physically interacted with Pad1, a component of the 26S proteasome (Figure 3c). This was further supported by experiments using temperature-sensitive proteasome mutant mts2 or mts3. Rad22 protein was stabilized and accumulated after inactivation of the proteasome machinery (Figure 3d).
Rad22 protein degradation by Bag101-mediated proteasome.
(a) The amount of Rad22 in cells overexpressing bag101 or containing bag101 deletions. Rad22 was measured with Western blots using anti-Rad22 antibodies. The Rad22 protein levels in no-irradiated cells were represented here and that of the time course after irradiation were shown in Supplementary Figure S2. (b) rad22 mRNA level in cells overexpressing or containing bag101 deletions. The mRNA from each mutant was measured with quantitative PCR using specific rad22 primers. hxk2 was used for internal controls. These experiments were performed three times and the error bars represent the S.D. (c) Physical interaction of the 26S proteasome with Bag101. Pad1 was co-immunoprecipitated with Bag101 in cell lysates expressing Bag101-FH6 and immunoblotted with anti-Pad1 antibody. (d) The stabilization of Rad22 proteins after proteasome inactivation. Rad22 was highly expressed in temperature-sensitive mutants mts2 and mts3 compared to cells expressing wild type mts2 and mts3 after inactivating proteasomes by increasing the temperature from 26°C to 36°C.
Bag101 interacts with Rad22 through its BAG domain and controls Rad22-dependent HR
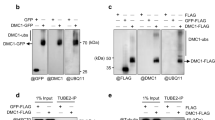
It has been reported that Bag101 binds to Hsc/Hsp70 through its BAG domain at the C-terminus and binds to the 26S proteasome through the UBL domain at the N-terminus27,28. Specific domain-deleted mutants of Bag101 were generated in order to determine which region was responsible for binding to Rad22 (Figure 4a). An immunoprecipitation experiment with FLAG antibodies from yeast cells expressing GFP-tagged Rad22 and FH6-tagged wild type or domain-deletion mutants of Bag101 revealed that Rad22 binds to the BAG domain of Bag101 (Figure 4b). The HR frequency and Rad22 focus formation in the presence of overexpression was examined in these BAG- and UBL-deleted mutants. Although overexpression of wild type bag101 significantly decreased both HR frequency and Rad22 focus formation, these were not affected by overexpression of either deletion mutant, suggesting a critical role for the interaction of Bag101 with both Rad22 and the 26S proteasome during Rad22 protein degradation and an association of this with the HR frequency (Figures 4c and 4d). This was supported by the observation that the Rad22 protein level was stabilized in both BAG- and UBL-deleted mutants (Figure 4e).
Regulation of HR by binding of Bag101 to Rad22 at the BAG domain.
(a) Illustration of Bag101 domains and the deletion mutants. (b) Physical interaction of Rad22 with Bag101 through the BAG domain. Immunoprecipitation was performed with FH6-tagged Bag101 for each cell containing a deleted domain; and immunoblotting was performed with anti-FLAG and anti-GFP antibodies to detect interactions. (c) HR frequency in each deletion mutant. Spontaneous HR frequencies were measured with the RDUX200 reporter gene. These experiments were performed three times and error bars represent the S.D. (d) Rad22-foci in deletion mutants. Cells were collected to count the number of Rad22-focus positive cells. Error bars represent the S.D. (e) The amount of Rad22 in Bag101 domain-deletion mutants. Rad22 levels in each mutant lacking the BAG or UBL domain were measured with Western blots using anti-Rad22 antibodies. (f) Dissociation of Rad22 from Bag101 after IR exposures. Cells were irradiated with 500 Gy and at the indicated times after irradiation, were harvested for immunoprecipitation with a HATA tag. Cell lysates and immunoprecipitates were immunoblotted with anti-Rad22 and anti-HA antibodies. The intensities of the Rad22 band were calibrated with a value of 1.0 with 0 Gy.
Finally, the role of Bag101 in Rad22 degradation following irradiation was investigated. Immunoprecipitation with Bag101-HATA revealed that, although significant amounts of Rad22 bound to Bag101, Rad22 began to dissociate from Bag101 immediately after irradiation and the Rad22-bound form of Bag101 disappeared 5 hours later. As expected from experiments with domain-deletion mutants of bag101, Rad22 protein levels increased with this increased dissociation (Figure 4f). Taken together, these results indicate that Bag101 regulates Rad22 protein levels through an interaction with its BAG domain and contributes to HR regulation after IR exposures.
Discussion
In this study, a novel regulatory mechanism was suggested for HR: this mechanism acts through Rad22 degradation and decreased Rad22 focus formation. Immunoprecipitation with Rad22-FLATA identified sixteen Rad22-binding proteins including Bag101. Bag101 suppressed Rad22 focus formation and HR activity both in non-irradiated and irradiated cells. Conversely, lack of Bag101 resulted in increased HR activity, thereby increasing cell viability after IR exposures. Experiments with domain-deletion mutants revealed that Bag101 binds to Rad22 through the BAG domain and a lack of this interaction increased Rad22 accumulation and consequently enhanced HR activity. Indeed, Rad22 dissociated from Bag101 immediately after IR exposures and accumulated significantly in irradiated cells.
Rad22 levels could be controlled by 26S proteasome-mediated degradation, since the level of Rad22 mRNA did not decrease (Figure 2b). This is supported by the observation that the amount of Rad22 protein increased greatly after inactivation of the 26S proteasome machinery in temperature sensitive mutants of mts2 and mts3. Consistent with this, it was found that there was an interaction between Bag101 and Pad1, a component of the 26S proteasome (Figure 3c). It has been reported that BAG-1 interacts with 26S proteasome components and also target proteins, such as the glucocorticoid hormone receptor and consequently regulates the degradation of misfolded proteins and affects the quality of cellular proteins27. This is the case for control of Rad22 protein levels in DNA repair pathways, where they should be correctly folded. The failure of this association of Rad22 with Bag101 led to the stabilization of Rad22 proteins and resulted in enhanced HR activity as observed in bag101-deletion mutants and in irradiated cells (Figures 3a and 4f). Sacher et al. proposed a model that SUMO modification of Rad22 sustains its activity by protecting it from proteasome-mediated degradation18. Moreover, the deficiency in SUMOylation significantly reduced intrachromosome recombination, although it marginally affects interchromosomal recombination. This is in agreement with the present results showing significantly decreased HR activity (intrachromosome recombination) in bag101 overexpressing cells through Rad22 degradation (Figures 1c and 2a). Therefore, Bag101 could a major contributor to HR regulation in the absence or presence of DNA damage, although modifications by other factors, such as SUMOylation, could affect the stability of Rad22.
BAG-1 and its family of proteins are conserved throughout eukaryotes, including vertebrates, insects, nematodes, yeast and plants29,30. Consequently it appears plausible that BAG proteins could be a crucial factor for the maintenance of genome integrity. Indeed, BAG family proteins are overexpressed in several tumors, such as leukemia, breast cancer, prostate cancer and colon cancers31,32,33. Although this study focused on Rad22, Bag101 might facilitate the degradation of other proteins involved in DSB repair. In fact, several DNA repair proteins, such as MDC1, BRCA1 and Ku80, have been reported to be modified by ubiquitination and subsequently destroyed through the proteasome pathway34,35,36,37. Thus, overexpression of BAG1 proteins in tumors might lead to deficient levels of DNA repair due to the presence of lower levels of repair proteins and this could eventually lead to the development of cancer. This is substantiated by the fact that repair-deficient hereditary diseases or repair protein-deficient mice have a predisposition to develop cancer38,39. These findings concerning Bag101 might help to provide a useful tool to study mechanistic links between HR and carcinogenesis and help to develop new therapeutic agents.
Methods
Yeast strains, plasmid construction and techniques
Yeast strains were manipulated using standard genetic techniques as previously described21,22,23. All yeast strains used in this study are listed in Supplementary Table S1. The transformant constitutively expressing the protein was generated by subcloning each cDNA into a pFOX1-based vector with TEF promoter. The plasmids or cDNA constructs were shown in Supplementary Table S2. The primers used in this study are listed in Supplementary Table S3. Yeast cells were grown in the appropriate medium: cells without overexpression strains were grown at 30°C in yeast extract medium (YE), while overexpression strains were grown in Edinburgh minimal medium (EMM) containing uracil for selection. The temperature-sensitive proteasomal mutants mts2 and mts3, which have mutations in 19S proteasome regulatory subunits rpt2 and rpt12 genes respectively, were grown at 26°C and shifted to 36°C for inactivation of the proteasome machinery.
HR frequency assay and survival assays
HR frequency assays were performed as previously reported22. Briefly, a recombination reporter cassette ura4::kanMX6 with a 200-bp tandem duplication (RDUX200(+)) was integrated into endogenous ura4+ locus, which confers uracil prototrophy with the concomitant loss of G-418 resistance upon HR of the duplication. Cells were cultured in YE medium containing 500 μg/ml of G418 overnight to eliminate recombinants. The cells were collected by centrifugation, washed once with sterilized water and then, resuspended in appropriate concentrations with sterilized water. HR frequencies were detected with colony formation on uracil-free tester plates at a density of 2–5 × 105 cells per plate where ura+ recombinants can form colonies. For determination of IR-induced HR frequencies, cells were irradiated with 250 and 500 Gy of γ-rays just before collection. Cell survival was determined with colony formation assays on YE plates at a density of 2–4 × 102 cells per plate.
Rad22-foci formation
GFP was integrated into the Rad22 allele and was used for assays of endogenous Rad22-foci formation. Cells were irradiated with 250 and 500 Gy of γ-rays and then cultured for 30 minutes. The cultures were cooled on ice with Hoechst 33342 for 30 minutes to stain nuclei. The cells were mounted on glass slides and sealed with nail polish. Rad22 foci formation was observed with a Nikon Eclipse 90i fluorescent microscope. The Rad22-focus positive cells were defined as more than one foci-containing cells. The experiments were performed at least twice and more than 100 cells were counted in each experiment.
Immunoprecipitation
For purification of Rad22-binding proteins, Rad22 was tagged at the C-terminus with FLATA (FLAG - TEV cutting site – Protein A). To confirm the interaction between Rad22 and the proteins screened with LC-MS/MS analysis, reverse immunoprecipitation was carried out with FH6 (FLAG - 6× His) tagged proteins. To investigate the interaction of Rad22 and Bag101 following irradiation, HATA (HA - TEV cutting site - ProteinA) was tagged on the C-terminus of Bag101. Cells expressing each construct were suspended in lysis buffer (50 mM HEPES, pH 7.6, 300 mM potassium acetate, 5 mM magnesium acetate, 20 mM β-glycerol phosphate, 1 mM EGTA, 1 mM EDTA, 0.1% (v/v) Nonidet P-40), chilled in liquid nitrogen and homogenized. After removal of debris with centrifugation, the beads were added to samples and rotated at 4°C for 1 hour: human IgG Sepharose (GE healthcare) was used for FLATA tag and HATA tag, or an anti-FLAG M2 Affinity Gel (SIGMA-Aldrich) for a FH6 tag. The precipitates were washed with wash buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 0.25% (v/v) NP-40, 5% (v/v) glycerol) and treated with TEV protease at room temperature for 1 hour. For a FLATA tag, anti-FLAG M2 Affinity Gels were added to the supernatants and were eluted with FLAG peptides. The precipitated proteins were separated on polyacrylamide gels and analyzed with a LC-MS/MS mass spectrometer.
Immunoblotting
The cells were harvested with centrifugation, resuspended in 1 M NaOH and then placed at room temperature for 10 minutes. The precipitates obtained from centrifugation were resuspended in 2× sample buffer (0.1 M Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 0.1 M DTT) and heated at 100°C for 5 minutes. After the removal of debris with centrifugation, the proteins were separated on a polyacrylamide gel and analyzed with immunoblotting using anti-Rad22 (BAM-63-003-EX, COSMO BIO), anti-beta Actin (ab8224, abcam), anti-FLAG M2 (F1804, SIGMA-Aldrich), anti-phospho-Histone H2A (Ser129) (07-745, upstate) and anti-HA high affinity (3F10, Roche Applied Science) antibodies. The chemiluminescent signals were measured with a LAS-3000 luminescent image analyzer (Fuji Film).
Quantification of Rad22 protein and mRNA
The quantity of Rad22 proteins was measured with immunoblotting. Western blots were scanned with a LAS-3000 luminescent image analyzer and the intensities of the Rad22 bands were measured using Multi Gauge ver.3.0 imaging software. The quantity of rad22 mRNA per cell was measured with quantitative PCR and hexokinase 2 was used for internal controls.
LC-MS/MS analysis
LC-MS/MS analysis was performed as reported previously24 and the direct nanoflow LC-MS system was equipped with a quadrupole-time-of-flight hybrid mass spectrometer (Q-Tof Ultima, Waters, Bedford, MA, USA). Database searches were performed using MASCOT software (version 2.3, Matrix Science Ltd., London)25.
References
Pâques, F. & Haber, J. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and molecular biology reviews 63, 349–404 (1999).
Melo, J. & Toczyski, D. A unified view of the DNA-damage checkpoint. Current Opinion in Cell Biology 14, 237–245 (2002).
Wyman, C. & Kanaar, R. DNA double-strand break repair: all's well that ends well. Annual review of genetics 40, 363–83 (2006).
Friedl, a. a., Kiechle, M., Fellerhoff, B. & Eckardt-Schupp, F. Radiation-induced chromosome aberrations in Saccharomyces cerevisiae: influence of DNA repair pathways. Genetics 148, 975–88 (1998).
Krogh, B. O. & Symington, L. S. Recombination proteins in yeast. Annual review of genetics 38, 233–71 (2004).
Symington, L. S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiology and molecular biology reviews 66, 630–670 (2002).
Shinohara, a., Shinohara, M., Ohta, T., Matsuda, S. & Ogawa, T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes to cells: devoted to molecular & cellular mechanisms 3, 145–56 (1998).
Mortensen, U. H., Bendixen, C., Sunjevaric, I. & Rothstein, R. DNA strand annealing is promoted by the yeast Rad52 protein. Proceedings of the National Academy of Sciences of the United States of America 93, 10729–34 (1996).
Hays, S. & Firmenich, A. Studies of the Interaction between Rad52 Protein and the Yeast Single-Stranded DNA Binding Protein RPA. Molecular and cellular biology 18, 4400–06 (1998).
Sung, P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. Journal of Biological Chemistry 272, 28194–28197 (1997).
New, J., Sugiyama, T., Zaitseva, E. & Kowalczykowski, S. C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410 (1998).
Shinohara, A., Ogawa, H. & Ogawa, T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69, 457–70 (1992).
Watt, P., Hickson, I., Borts, R. & Louis, E. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144, 935–45 (1996).
Sinclair, D. a. Accelerated Aging and Nucleolar Fragmentation in Yeast sgs1 Mutants. Science 277, 1313–1316 (1997).
Pfander, B., Moldovan, G.-L., Sacher, M., Hoege, C. & Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–33 (2005).
Krejci, L. et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–09 (2003).
Veaute, X., Jeusset, J. & Soustelle, C. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–12 (2003).
Sacher, M., Pfander, B., Hoege, C. & Jentsch, S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nature cell biology 8, 1284–90 (2006).
Ohuchi, T. et al. Rad52 sumoylation and its involvement in the efficient induction of homologous recombination. DNA repair 7, 879–89 (2008).
Altmannova, V. et al. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic acids research 38, 4708–21 (2010).
Suga, M. & Hatakeyama, T. A rapid and simple procedure for high-efficiency lithium acetate transformation of cryopreserved Schizosaccharomyces pombe cells. Yeast (Chichester, England) 22, 799–804 (2005).
Takeda, J. et al. Radiation induction of delayed recombination in Schizosaccharomyces pombe. DNA repair 7, 1250–61 (2008).
Sato, M., Dhut, S. & Toda, T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast (Chichester, England) 22, 583–91 (2005).
Ichimura, T. et al. Phosphorylation-dependent interaction of kinesin light chain 2 and the 14-3-3 protein. Biochemistry 41, 5566–72 (2002).
Kakiuchi, K. et al. Proteomic analysis of in vivo 14-3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry 46, 7781–92 (2007).
Lüders, J., Demand, J. & Höhfeld, J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. The Journal of biological chemistry 275, 4613–7 (2000).
Demand, J., Alberti, S., Patterson, C. & Höhfeld, J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Current biology: CB 11, 1569–77 (2001).
Briknarová, K. et al. BAG4/SODD protein contains a short BAG domain. The Journal of biological chemistry 277, 31172–8 (2002).
Takayama, S., Xie, Z. & Reed, J. C. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. The Journal of biological chemistry 274, 781–6 (1999).
Sondermann, H. Structure of a Bag/Hsc70 Complex: Convergent Functional Evolution of Hsp70 Nucleotide Exchange Factors. Science 291, 1553–1557 (2001).
Takayama, S., Krajewski, S. & Krajewska, M. Expression and Location of Hsp70/Hsc-Binding Anti-Apoptotic Protein BAG-1 and Its Variants in Normal Tissues and Tumor Cell Lines. Cancer research 58, 3116–3131 (1998).
Brimmell, M. et al. High level expression of differentially localized BAG-1 isoforms in some oestrogen receptor-positive human breast cancers. British journal of cancer 81, 1042–51 (1999).
Turner, B. C. et al. BAG-1: a novel biomarker predicting long-term survival in early-stage breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 19, 992–1000 (2001).
Shi, W. et al. Disassembly of MDC1 foci is controlled by ubiquitin-proteasome-dependent degradation. The Journal of biological chemistry 283, 31608–16 (2008).
Choudhury, A. D., Xu, H. & Baer, R. Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. The Journal of biological chemistry 279, 33909–18 (2004).
Postow, L. et al. Ku80 removal from DNA through double strand break-induced ubiquitylation. The Journal of cell biology 182, 467–79 (2008).
Postow, L. Destroying the ring: Freeing DNA from Ku with ubiquitin. FEBS letters 585, 2876–2882 (2011).
Brodie, S. G. & Deng, C. X. BRCA1-associated tumorigenesis: what have we learned from knockout mice? Trends in genetics: TIG 17, S18–22 (2001).
Bouwman, P. & Jonkers, J. Mouse models for BRCA1 associated tumorigenesis. Cell Cycle 7, 2647–2653 (2008).
Acknowledgements
This work was supported in part by a Grant-in Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan and also by a cooperative research with Radiation Biology Center, Kyoto University.
Author information
Authors and Affiliations
Contributions
Y.S. performed most of the experiments, analyzed the data and co-wrote the manuscript. J.T., M.O. conceived the experiment. J.K., A.K., K.H., M.T. contributed to the data analyses and discussion. T.M. performed the experiment with mts mutants. K.K. co-wrote the manuscript. T.I. supervised the study.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Figures
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Saito, Y., Takeda, J., Okada, M. et al. The proteasome factor Bag101 binds to Rad22 and suppresses homologous recombination. Sci Rep 3, 2022 (2013). https://doi.org/10.1038/srep02022
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02022
This article is cited by
-
DNA replication machinery prevents Rad52-dependent single-strand annealing that leads to gross chromosomal rearrangements at centromeres
Communications Biology (2020)
-
UBL/BAG-domain co-chaperones cause cellular stress upon overexpression through constitutive activation of Hsf1
Cell Stress and Chaperones (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.