Abstract
Orchid epiphytes, a group containing at least 18,000 species, thrive in habitats that often undergo periodic drought stress. However, few global gene expression profiling datasets have been published for studies addressing the drought-resistant mechanism of this special population. In this study, an experiment involving the effect of continuous drought treatments on an epiphytic orchid, Dendrobium catenatum, was designed to generate 39 mature-leaf-tissue RNA-seq sequencing datasets with over two billion reads. These datasets were validated by a series of quality assessments including RNA sample quality, RNA-seq read quality, and global gene expression profiling. We believe that these comprehensive transcriptomic resources will allow a better understanding of the drought-resistant mechanisms of orchid epiphytes.
Design Type(s) | transcription profiling by high throughput sequencing design |
Measurement Type(s) | drought tolerance |
Technology Type(s) | RNA sequencing |
Factor Type(s) | temporal_interval • water content |
Sample Characteristic(s) | Dendrobium catenatum • leaf |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
In response to prolonged water deficit stress, plants have evolved coping mechanisms to increase their drought tolerance through physical adaptations, molecular regulations, and environmentally suitable metabolic pathways1–3. Most studies concerning drought stress mechanisms have been performed in Arabidopsis thaliana and other drought-intolerant C3 plants2. Studying a highly resistant plant that has been shaped by natural selection is the most direct and effective way to extract crucial genes and determine the main metabolic pathways of the drought stress procedure.
In the wild, most epiphytic orchids, a prosperous group containing over 18,000 species, take root on the surface of tree bark or rocks4,5. Due to the poor moisture supply in these habitats6, these plants usually suffer periodic water shortage7. While adapting to harsh habitats, some orchid species have evolved succulent storage-organs, such as pseudobulbs8,9, thick leaves10, and crassulacean acid metabolism (CAM)11, a photosynthetic pathway with high water-use efficiency12. Morphological and anatomical studies show that orchid plants possess desirable qualities for mitigating drought stress10,13,14. By measuring physiological indexes and secondary metabolites of Dendrobium moniliforme15, Wu et al. found that increasing antioxidant enzyme activities and osmolytes play an important role in protecting plants under drought stress. Although several physiological traits might provide clues for the mechanism of drought resistance, there is no large data set that allows holistic understanding. Unfortunately, few comprehensive transcriptomic profiling studies that address drought resistance have been published.
Comparing the two published genomes from epiphytic orchid species16,17, Phalaenopsis equestris and Dendrobium catenatum, the latter possesses more Heat-shock protein 70 family members and R genes17, which suggests that D. catenatum can tolerate a much wider variety of environments and has superior qualities for adverse resistance. A previous study demonstrates that D. catenatum uses the facultative CAM pathway as a drought-enduring process11. Hence, this species can be considered as drought-resistant material useful for elucidating mechanisms of mitigating drought stress in epiphytic orchids. Previous studies show that the circadian clock modifies responsiveness to environmental input and stress according to the time of day18–21. With regard to the correlation between CAM and circadian rhythm22. the conventional sampling tactics that focus on a single time point per day should be abandoned as, if the daylight sampling time is fixed, some important clues to key resistance genes could be missed.
In the current study, D. catenatum plants were subjected to continuous drought treatments by simulating their natural environment under controlled conditions. Sampling time points were set for both day and night during the drought procedure. A dataset containing 39 RNA-seq with over 41 million sequence reads per sample was generated using the Illumina HiSeq 2500 platform. We assessed RNA sample quality, RNA-seq read quality, and the global gene profile (Fig. 1) to ensure the dependability of our dataset. We believe that these transcriptomic profiles will contribute to a comprehensive understanding of the mechanism of drought resistance in D. catenatum.
The raw data were filtered using the package Fastq_clean, and clean data were assessed using FastQC and MultiQC. The clean reads were mapped to the D. catenatum genome (GenBank Assembely ID ASM160598v2) using Hisat2. The package ReSQC was used to calculate RNA-seq reads coverage over the gene body. Gene abundance was quantified using DESeq2.
Methods
Plant material and experimental design
Clones of D. catenatum were planted in transparent plastic pots (5.0 cm in diameter) with sphagnum moss as the matrix. Eight-month-old plants were transferred into a phytotron chamber (12/12 h light/dark, light intensity ~100 μmol m−2s−1; 28/22 °C day/night; and relative humidity 60/70% day/night) and adapted to the controlled conditions for 10 days before being used for the follow-up experiment. The experiments were conducted on initially healthy individuals (~12 cm height). Plants were irrigated on the first day and then water was withheld to mimic drought stress. We collected leaf samples when the volumetric water content of the base material declined to ~30–35%, ~10–15, and ~0%, respectively, at both 09:00 h and 21:00 h (Fig. 1). The fourth and fifth leaves (mature leaf) from the apex of each plant were harvested and mixed to create one sample. These samples were immediately frozen in liquid N2 and stored at −80 °C.
RNA isolation and sequencing
Total RNA was extracted from the samples mentioned above (Table 1) using the RNAprep Pure Plant Kit (No. DP441; Polysaccharides & Polyphenolics-rich; Tiangen Co. Ltd, Beijing, China; http://www.tiangen.com/) according to the manufacturer’s protocols. RNA purity was estimated using a NanoPhotometer® spectrophotometer (Implen, CA, USA). RNA quality was assessed using an RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). RNA samples of acceptable quality were used to construct non-strand-specific sequencing libraries with the TruSeq RNA Sample Prep Kit (Illumina, CA, USA). These libraries were sequenced using the PE150 mode on an Illumina HiSeq2500 platform at Novogene Corporation (Beijing, China; http://www.novogene.com/).
Data filtering and assessment
The raw data (raw reads; Data Citation 1) were filtered using Fastq_clean v2.023. Sequencing adapters, low-quality bases, viral sequences, and rRNA sequences were cleaned. The criteria for this filtering procedure were set as follows: (1) RNA 5′ and 3′ adapters were set as [5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′] and [5′-GATCGGAAGAGCACACGTCTGAACTCCAGTCAC (index) ATCTCGTATGCCGTCTTCTGCTTG-3’] (the indexes are listed in Table 1), respectively; (2) bases with a phred quality score below 20 were clipped from both ends of reads; (3) after low-quality bases were trimmed, reads containing over two “N” were discarded; (4) reads with a length shorter than 75 nt were discarded; and (5) the parameters for BWA v0.5.724 were set as recommended according to Fastq_clean instructions. The statistics of clean reads are listed in Table 1. The quality of the clean data was evaluated using the package FastQC v0.11.7 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and then summarized using MultiQC v1.325.
Gene quantification and detection of read coverage skewness
The clean reads were mapped to the D. catenatum genome17 (GenBank Assembly ID ASM160598v2) using Hisat226 with default parameters. Salmon v0.9.127 was used to estimate gene abundance as read counts in the alignment-based mode. The raw read counts were imported into the R package DESeq228 for normalization. We used the package ReSQC29 to assess RNA-seq read coverage skewness over the gene body based on the above mapping results.
Assessment of sample composition
A heatmap for cluster relationships among samples representing Poisson distance were generated with raw read counts. The R package PoiClaClu30 was used for the calculation of Poisson distance, and the R package Pheatmap (https://cran.r-project.org/web/packages/pheatmap/index.html) for visualization. A principal component analysis (PCA) was also employed to assess sample relationships based on rlog-transformed values of raw read counts.
Gene hierarchical clustering and Gene Ontology (GO) analysis
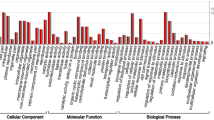
To determine the highly correlated genes in this prolonged drought experiment, weighted gene co-expression network analysis (WGCNA)31 was used to detected gene clusters (modules) on normalized read counts (Data Citation 2) using the WGCNA v1.6332,33 package in R. This analysis generated a topological overlap matrix plot (Fig. 2) that illustrated the relationships among gene clusters. To give an insight into the functions of both genes and gene clusters, we performed GO enrichment analysis using Gogsea, a web tool from Omicshare (http://www.omicshare.com/tools/Home/Soft/gogsea). The edge information of each gene cluster and the results of both GO annotation and GO enrichment are stored in the Figshare repository (Data Citation 3).
Code availability
The R scripts for reads count filtration and normalization, heatmap illustration, PCA and WGCNA are available in Figshare (Data Citation 4).
Data Records
The RNA-seq raw data of 39 samples are deposited at the NCBI Sequence Read Archive (Data Citation 1).
Supplementary materials are available on the Figshare data management platform (Data Citations 2, 3, 4). Data Citation 2 provides expression profiles of raw read counts and normalized read counts; Data Citation 3 contains WGCNA results, GO annotation for all genes, and GO enrichment for gene clusters. Data Citation 4 is dedicated to the R scripts in this study.
Technical Validation
RNA quality control
The quality of total RNA is a critical parameter for the construction of sequencing libraries and the follow-up quantitative analyses. In particular, RNA integrity (RIN) is positively correlated on uniquely mapped reads in RNA-Seq34, which means low RIN would lead to a bias in gene expression profiles. In this study, RNA samples with a RIN value >6.5 were employed for RNA-seq library construction, which meant that high-quality reads were obtained for subsequent studies. The quality values for RNA samples, including RIN, are listed in Table 2.
Quality validation
The high clean data rate (Table 1), ranging from 98.73% to 99.56%, indicated that both RNA-seq libraries and raw RNA-seq data obtained in this study were of high quality. Results of clean reads assessment by FastQC are illustrated in Fig. 3. The per base quality scores were >30, and most per sequence quality scores were >20, suggesting a high sequence quality. The per sequence GC contents had pattern curves similar to a normal distribution indicating the sequencing data were free of contamination. In addition, we examined read-mapping qualities of the 39 samples, including mapping rates and read distribution on reference genes. The mapping rates to the reference genome were superior, with a range from 83.36% to 88.02% (Table 1). The distribution of reads based on the detection of read coverage skewness showed good fragmentation randomness (Fig. 4), which reflected that each part of the gene was sequenced evenly.
Both the heatmap (Fig. 5a) and PCA (Fig. 5b) of gene profiles from all 39 samples revealed the clustering of samples according to time and drought level. The samples from daytime and nighttime clustered into two separate groups. The extreme drought groups during both day and night were distinctly separate from the groups with water content of 10–15% and 25–30%. However, the clustering of samples with 10–15% and 25–30% water content overlapped. The explanation for this is that, for a CAM plant, moderate drought would not result in a significant change in gene expression because of its strong ability to adapt to drought.
Additional information
How to cite this article: Wan, X. et al. Transcriptomic profiling for prolonged drought in Dendrobium catenatum. Sci. Data. 5:180233 doi: 10.1038/sdata.2018.233 (2018).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
References
Chaves, M. M., Maroco, J. P. & Pereira, J. S. Understanding plant responses to drought - From genes to the whole plant. Funct. Plant Biol. 30, 239–264 (2003).
Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 167, 313–324 (2016).
Khanna-Chopra, R . & Singh, K. Drought resistance in crops: physiological and genetic basis of traits for crop productivity. Stress Responses in Plants (eds Tripathi, B. N. & Müller, M.) 267–292 (Springer, 2015).
Dressler, R. L. . Phylogeny and Classification of the Orchid Family. (Cambridge University Press, 1993).
Atwood, J. The size of Orchidaceae and the systematic distribution of epiphytic orchids. Selbyana 9, 171–186 (1986).
Rapp, J. M. & Silman, M. R. Epiphyte response to drought and experimental warming in an Andean cloud forest. F1000Research 3, 1–29 (2014).
Zotz, G. & Winkler, U. Aerial roots of epiphytic orchids: The velamen radicum and its role in water and nutrient uptake. Oecologia 171, 733–741 (2013).
Ng, C. K. Y. & Hew, C. S. Orchid pseudobulbs - ‘False’ bulbs with a genuine importance in orchid growth and survival!. Scientia Horticulturae 83, 165–172 (2000).
Zimmerman, J. Role of pseudobulbs in growth and flowering of Catasetum viridiflavum (Orchidaceae). Am. J. Bot. 77, 533–542 (1990).
Zhang, S.-B. et al. Differentiation of water-related traits in terrestrial and epiphytic Cymbidium species. Front. Plant Sci. 6, 1–10 (2015).
Zhang, Z., Dongxian, H. & Rongfu, G. Concomitant CAM and C3 photosynthetic pathways in Dendrobium officinale plants. J. Am. Soc. Hortic. Sci. 139, 290–298 (2014).
Ahmad, P. . Water Stress and Crop Plants: A Sustainable Approach. (Wiley Blackwell Press, 2016).
Mollenhauer, H. H. A Comparison of root cap cells of epiphytic, Terrestrial and Aquatic Plants. Am. J. Bot. 54, 1249 (1967).
Rodrigues, M. A. et al. Spatial patterns of photosynthesis in thin- and thick-leaved epiphytic orchids: Unravelling C3-CAM plasticity in an organ-compartmented way. Ann. Bot 112, 17–29 (2013).
Wu, X., Yuan, J., Luo, A., Chen, Y. & Fan, Y. Drought stress and re-watering increase secondary metabolites and enzyme activity in Dendrobium moniliforme. Ind. Crops Prod 94, 385–393 (2016).
Liu, Z. J. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47, 65–72 (2015).
Zhang, G. Q. et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep 6, 1–10 (2016).
Greenham, K. & McClung, C. R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16, 598–610 (2015).
Greenham, K. et al. Temporal network analysis identifies early physiological and transcriptomic indicators of mild drought in Brassica rapa. Elife 6, 1–26 (2017).
Legnaioli, T., Cuevas, J. & Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 28, 3745–3757 (2009).
Yakir, E., Hilman, D., Harir, Y. & Green, R. M. Regulation of output from the plant circadian clock. FEBS J 274, 335–345 (2007).
Smith, J. A. C. & Winter, K. Crassulacean acid metabolism : a continuous or discrete trait ? New Phytol. 208, 73–78 (2015).
Zhang, M., Zhan, F., Sun, H. & Gong, X. Fastq_clean, An optimized pipeline to clean the Illumina sequencing data with quality control. In IEEE International Conference on Bioinformatics and Biomedicine 44–48 (IEEE, 2014).
Li, H. & Richard, D. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wang, L., Wang, S. & Li, W. RSeQC : quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012).
Witten, D. M. Classification and clustering of sequencing data using a poisson model. Ann. Appl. Stat 5, 2493–2518 (2011).
Zhang, B. & Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article 17 (2005).
Langfelder, P. & Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Langfelder, P. & Horvath, S. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 46, 1–17 (2012).
Chen, E. A. et al. Effect of RNA integrity on uniquely mapped reads in RNA-Seq. BMC Res. Notes 7, 753 (2014).
Data Citations
Wan, X., Zou, L.-H., Zheng, B.-Q., Tian, Y.-Q., & Wang, Y. NBCI Sequence Read Archive SRP132541 (2018)
Wan, X., Zou, L.-H., Zheng, B.-Q., Tian, Y.-Q., & Wang, Y. Figshare https://doi.org/10.6084/m9.figshare.6959924 (2018)
Wan, X., Zou, L.-H., Zheng, B.-Q., Tian, Y.-Q., & Wang, Y. Figshare https://doi.org/10.6084/m9.figshare.6960377 (2018)
Wan, X., Zou, L.-H., Zheng, B.-Q., Tian, Y.-Q., & Wang, Y. Figshare https://doi.org/10.6084/m9.figshare.6954590 (2018)
Acknowledgements
This work was supported by the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (Grant No. 2013BAD01B0703; The Exploitation, Innovation and Utilization of Genus Dendrobium Germplasm Resources).
Author information
Authors and Affiliations
Contributions
X.W. designed the experiments, collected samples, extracted RNA, processed the data, and wrote the original draft. L.-H.Z. designed the experiments, collected samples, extracted RNA, processed the data, and wrote the original draft. B.-Q.Z. collected samples, extracted RNA, and reviewed the manuscript. Y.-Q.T. supplied the original clones of Dendrobium catenatum, and reviewed the manuscript. Y.W. designed the experiments, reviewed the manuscript and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
ISA-Tab metadata
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files made available in this article.
About this article
Cite this article
Wan, X., Zou, LH., Zheng, BQ. et al. Transcriptomic profiling for prolonged drought in Dendrobium catenatum. Sci Data 5, 180233 (2018). https://doi.org/10.1038/sdata.2018.233
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/sdata.2018.233