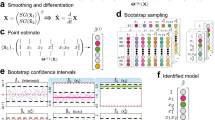
Abstract
All physical laws are described as mathematical relationships between state variables. These variables give a complete and non-redundant description of the relevant system. However, despite the prevalence of computing power and artificial intelligence, the process of identifying the hidden state variables themselves has resisted automation. Most data-driven methods for modelling physical phenomena still rely on the assumption that the relevant state variables are already known. A longstanding question is whether it is possible to identify state variables from only high-dimensional observational data. Here we propose a principle for determining how many state variables an observed system is likely to have, and what these variables might be. We demonstrate the effectiveness of this approach using video recordings of a variety of physical dynamical systems, ranging from elastic double pendulums to fire flames. Without any prior knowledge of the underlying physics, our algorithm discovers the intrinsic dimension of the observed dynamics and identifies candidate sets of state variables.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Code availability
The open-source code to reproduce our training and evaluation results is available at the Zenodo repository52 and GitHub (https://github.com/BoyuanChen/neural-state-variables).
References
Anderson, P. W. More is different. Science 177, 393–396 (1972).
Thompson, J. M. T. & Stewart, H. B. Nonlinear Dynamics and Chaos (Wiley, 2002).
Hirsch, M. W., Smale, S. & Devaney, R. L. Differential Equations, Dynamical Systems, and an Introduction to Chaos (Academic, 2012).
Kutz, J. N., Brunton, S. L., Brunton, B. W. & Proctor, J. L. Dynamic Mode Decomposition: Data-Driven Modeling of Complex Systems (SIAM, 2016).
Evans, J. & Rzhetsky, A. Machine science. Science 329, 399–400 (2010).
Fortunato, S. et al. Science of science. Science 359, eaao0185 (2018).
Bongard, J. & Lipson, H. Automated reverse engineering of nonlinear dynamical systems. Proc. Natl Acad. Sci. USA 104, 9943–9948 (2007).
Schmidt, M. & Lipson, H. Distilling free-form natural laws from experimental data. Science 324, 81–85 (2009).
King, R. D., Muggleton, S. H., Srinivasan, A. & Sternberg, M. Structure–activity relationships derived by machine learning: the use of atoms and their bond connectivities to predict mutagenicity by inductive logic programming. Proc. Natl Acad. Sci. USA 93, 438–442 (1996).
Waltz, D. & Buchanan, B. G. Automating science. Science 324, 43–44 (2009).
King, R. D. et al. The robot scientist Adam. Computer 42, 46–54 (2009).
Langley, P. BACON: a production system that discovers empirical laws. In Proc. Fifth International Joint Conference on Artificial Intelligence Vol. 1 344 (Morgan Kaufmann, 1977).
Langley, P. Rediscovering physics with BACON.3. In Proc. Sixth International Joint Conference on Artificial Intelligence Vol. 1 505–507 (Morgan Kaufmann, 1979).
Crutchfield, J. P. & McNamara, B. Equations of motion from a data series. Complex Syst. 1, 417–452 (1987).
Kevrekidis, I. G. et al. Equation-free, coarse-grained multiscale computation: enabling microscopic simulators to perform system-level analysis. Commun. Math. Sci. 1, 715–762 (2003).
Yao, C. & Bollt, E. M. Modeling and nonlinear parameter estimation with Kronecker product representation for coupled oscillators and spatiotemporal systems. Physica D 227, 78–99 (2007).
Rowley, C. W., Mezić, I., Bagheri, S., Schlatter, P. & Henningson, D. S. Spectral analysis of nonlinear flows. J. Fluid Mech. 641, 115–127 (2009).
Schmidt, M. D. et al. Automated refinement and inference of analytical models for metabolic networks. Phys. Biol. 8, 055011 (2011).
Sugihara, G. et al. Detecting causality in complex ecosystems. Science 338, 496–500 (2012).
Ye, H. et al. Equation-free mechanistic ecosystem forecasting using empirical dynamic modeling. Proc. Natl Acad. Sci. USA 112, E1569–E1576 (2015).
Daniels, B. C. & Nemenman, I. Automated adaptive inference of phenomenological dynamical models. Nat. Commun. 6, 8133 (2015).
Daniels, B. C. & Nemenman, I. Efficient inference of parsimonious phenomenological models of cellular dynamics using S-systems and alternating regression. PloS ONE 10, e0119821 (2015).
Benner, P., Gugercin, S. & Willcox, K. A survey of projection-based model reduction methods for parametric dynamical systems. SIAM Rev. 57, 483–531 (2015).
Brunton, S. L., Proctor, J. L. & Kutz, J. N. Discovering governing equations from data by sparse identification of nonlinear dynamical systems. Proc. Natl Acad. Sci. USA 113, 3932–3937 (2016).
Rudy, S. H., Brunton, S. L., Proctor, J. L. & Kutz, J. N. Data-driven discovery of partial differential equations. Sci. Adv. 3, e1602614 (2017).
Udrescu, S.-M. & Tegmark, M. AI Feynman: a physics-inspired method for symbolic regression. Sci. Adv. 6, eaay2631 (2020).
Mrowca D. et al. Flexible neural representation for physics prediction. In Advances in Neural Information Processing Systems Vol. 31 (eds Bengio, S. et al.) (Curran Associates, 2018).
Champion, K., Lusch, B., Kutz, J. N. & Brunton, S. L. Data-driven discovery of coordinates and governing equations. Proc. Natl Acad. Sci. USA 116, 22445–22451 (2019).
Baldi, P. & Hornik, K. Neural networks and principal component analysis: learning from examples without local minima. Neural Netw. 2, 53–58 (1989).
Hinton, G. E. & Zemel, R. S. Autoencoders, minimum description length, and Helmholtz free energy. Adv. Neural Inf. Process. Syst. 6, 3 (1994).
Masci, J., Meier, U., Cireşan, D. & Schmidhuber, J. Stacked convolutional autoencoders for hierarchical feature extraction. In International Conference on Artificial Neural Networks 52–59 (Springer, 2011).
Bishop C. M. et al. Neural Networks for Pattern Recognition (Oxford Univ. Press, 1995).
Camastra, F. & Staiano, A. Intrinsic dimension estimation: advances and open problems. Inf. Sci. 328, 26–41 (2016).
Campadelli, P., Casiraghi, E., Ceruti, C. & Rozza, A. Intrinsic dimension estimation: relevant techniques and a benchmark framework. Math. Probl. Eng. 2015, 759567 (2015).
Levina, E. & Bickel, P. J. Maximum likelihood estimation of intrinsic dimension. In Proc. 17th International Conference on Neural Information Processing Systems 777–784 (MIT Press, 2005).
Rozza, A., Lombardi, G., Ceruti, C., Casiraghi, E. & Campadelli, P. Novel high intrinsic dimensionality estimators. Mach. Learn. 89, 37–65 (2012).
Ceruti, C. et al. DANCo: an intrinsic dimensionality estimator exploiting angle and norm concentration. Pattern Recognit. 47, 2569–2581 (2014).
Hein, M. & Audibert, J.-Y. Intrinsic dimensionality estimation of submanifolds in Rd. In Proc. 22nd International Conference on Machine Learning 289–296 (Association for Computing Machinery, 2005).
Grassberger, P. & Procaccia, I. in The Theory of Chaotic Attractors 170–189 (Springer, 2004).
Pukrittayakamee, A. et al. Simultaneous fitting of a potential-energy surface and its corresponding force fields using feedforward neural networks. J. Chem. Phys. 130, 134101 (2009).
Wu, J., Lim, J. J., Zhang, H., Tenenbaum, J. B. & Freeman, W. T. Physics 101: Learning physical object properties from unlabeled videos. In Proc. British Machine Vision Conference (BMVC) (eds Wilson, R. C. et al.) 39.1-39.12 (BMVA Press, 2016).
Chmiela, S. et al. Machine learning of accurate energy-conserving molecular force fields. Sci. Adv. 3, e1603015 (2017).
Schütt, K. T., Arbabzadah, F., Chmiela, S., Müller, K. R. & Tkatchenko, A. Quantum-chemical insights from deep tensor neural networks. Nat. Commun. 8, 13890 (2017).
Smith, J. S., Isayev, O. & Roitberg, A. E. ANI-1: an extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci. 8, 3192–3203 (2017).
Lutter, M., Ritter, C. & Peters, J. Deep Lagrangian networks: using physics as model prior for deep learning. In International Conference on Learning Representations (2019).
Bondesan, R. & Lamacraft, A. Learning symmetries of classical integrable systems. Preprint at https://arxiv.org/abs/1906.04645 (2019).
Greydanus, S. J., Dzumba, M. & Yosinski, J. Hamiltonian neural networks. Preprint at https://arxiv.org/abs/1906.01563 (2019).
Swischuk, R., Kramer, B., Huang, C. & Willcox, K. Learning physics-based reduced-order models for a single-injector combustion process. AIAA J. 58, 2658–2672 (2020).
Lange, H., Brunton, S. L. & Kutz, J. N. From Fourier to Koopman: spectral methods for long-term time series prediction. J. Mach. Learn. Res. 22, 1–38 (2021).
Mallen, A., Lange, H. & Kutz, J. N. Deep probabilistic Koopman: long-term time-series forecasting under periodic uncertainties. Preprint at https://arxiv.org/abs/2106.06033 (2021).
Chen B. et al. Dataset for the paper titled Discovering State Variables Hidden in Experimental Data (1.0). Zenodo https://doi.org/10.5281/zenodo.6653856 (2022).
Chen B. et al. BoyuanChen/neural-state-variables: (v1.0). Zenodo https://doi.org/10.5281/zenodo.6629185 (2022).
Acknowledgements
This research was supported in part by NSF AI Institute for Dynamical Systems grant 2112085 (to H.L.), DARPA MTO Lifelong Learning Machines (L2M) Program HR0011-18-2-0020 (to H.L.), NSF NRI grant 1925157 (to H.L.), NSF DMS grant 1937254 (to Q.D.), NSF DMS grant 2012562 (to Q.D.), NSF CCF grant 1704833 (to Q.D.), DE grant SC0022317 (to Q.D.) and DOE ASCR DE grant SC0022317 (to Q.D.).
Author information
Authors and Affiliations
Contributions
B.C. and H.L. proposed the research; B.C., K.H., H.L. and Q.D. performed experiments and numerical analysis, B.C. and K.H. designed the algorithms; B.C., K.H., I.C. and S.R. collected the dataset; B.C., K.H., H.L. and Q.D. wrote the paper; all authors provided feedback.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Bryan Daniels and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Jie Pan, in collaboration with the Nature Computational Science team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 What state variables describe these dynamical systems?.
What state variables describe these dynamical systems? Identifying state variables from raw observation data is a precursor step to discovering physical laws. The key challenge is to figure out how many variables will give a complete and non-redundant description of the system’s states, what are the candidate variables, and how the variables are dependent on each other. Our work studies how to retrieve possible set of state variables from data distributions non-linearly embedded in the ambient space.
Extended Data Fig. 2 PCA and Neural State Variables visualization.
PCA and Neural State Variables visualization. Here we visualize the interesting symmetrical structures encoded in the Neural State Variables from single pendulum (A) and rigid double pendulum (B) after applying PCA algorithm on them. The colors represent the value of different physical variables. The x-axis and y-axis represent different components of the Neural State Variables.
Supplementary information
Supplementary Information
Supplementary Figs. 1–12, Discussion and Tables 1–10.
Supplementary Video 1
Overview video.
Supplementary Video 2
ID estimation results.
Supplementary Video 3
Long-term prediction stability results.
Supplementary Video 4
Robust prediction results for the single-pendulum system.
Supplementary Video 5
Robust prediction results for the double-pendulum system.
Source data
Source Data Fig. 3
Source data to reproduce the figure results.
Source Data Fig. 4
Source data to reproduce the figure results.
Source Data Fig. 5
Source data to reproduce the figure results.
Source Data Fig. 6
Source data to reproduce the figure results.
Source Data Extended Data Fig. 2
Source data to produce Extended Data Fig. 2.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, B., Huang, K., Raghupathi, S. et al. Automated discovery of fundamental variables hidden in experimental data. Nat Comput Sci 2, 433–442 (2022). https://doi.org/10.1038/s43588-022-00281-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-022-00281-6
This article is cited by
-
Generative learning for nonlinear dynamics
Nature Reviews Physics (2024)
-
Incorporating physics into data-driven computer vision
Nature Machine Intelligence (2023)
-
Knowledge-integrated machine learning for materials: lessons from gameplaying and robotics
Nature Reviews Materials (2023)
-
Constructing custom thermodynamics using deep learning
Nature Computational Science (2023)
-
Using artificial intelligence to transform astrobiology
Nature Astronomy (2023)