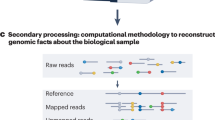
Abstract
Modern DNA sequencing is used as a readout for diverse assays, with the count of aligned sequences (read depth) representing the quantitative signal for each underlying cellular phenomena. Existing data formats for quantitative genomics assays are, however, limited in either the analysis speeds they enable, the disk space they require or both. We have developed the dense depth data dump (D4) format and tool suite, with the goal of balancing improved analysis speeds with file size. The D4 format is adaptive in that it profiles a random sample of aligned sequence depth from the input sequence file to determine an optimal encoding that enables fast data access. We demonstrate that the D4 format offers substantial speed improvements over existing formats for random access, aggregation and summarization, while also achieving better or comparable file sizes. This performance enables scalable downstream analyses that would be otherwise difficult.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The 1000 Genomes data were downloaded from http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/working/20190425_NYGC_GATK/. Bulk RNA-seq samples were downloaded from the ENCODE project website (www.encode.org) and a list of URLs to download all samples used is available at the D4utils GitHub repository (https://github.com/38/d4-format). The WGS dataset is sample HG002 from the Genome in a Bottle Project, and the RNA-seq dataset is sample ENCFF976QSN from the ENCODE project16. Source data are provided with this paper.
Code availability
The code underlying the software described and the data analyzed in this study are available at: D4utils, https://github.com/38/d4-format; D4 Rust API, https://docs.rs/d4; D4 Python API, https://github.com/38/pyd4. Code has also been deposited on Zenodo17 at https://zenodo.org/record/4684595#.YJQ272ZKidZ.
Change history
15 February 2022
A Correction to this paper has been published: https://doi.org/10.1038/s43588-022-00211-6
References
Sasani, T. A. et al. Large, three-generation human families reveal post-zygotic mosaicism and variability in germline mutation accumulation. Elife 8, e46922 (2019).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
2 Chromatin patterns at transcription factor binding sites. Nature https://doi.org/10.1038/nature28171 (2019).
Pedersen, B. S., Collins, R. L., Talkowski, M. E. & Quinlan, A. R. Indexcov: fast coverage quality control for whole-genome sequencing. Gigascience 6, 1–6 (2017).
Kent, W. J., Zweig, A. S., Barber, G., Hinrichs, A. S. & Karolchik, D. bigWig and bigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 (2010).
Frequently asked questions: data file formats. Genome Browser https://genome.ucsc.edu/FAQ/FAQformat.html (2021).
Koranne, S. Handbook of Open Source Tools 191–200 (Springer, 2011); https://doi.org/10.1007/978-1-4419-7719-9_10
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Fritz, M. H.-Y., Leinonen, R., Cochrane, G. & Birney, E. Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res. 21, 734–740 (2011).
Shao, Z., Reppy, J. H. & Appel, A. W. Unrolling lists. SIGPLAN Lisp Pointers VII, 185–195 (1994).
Pedersen, B. S. & Quinlan, A. R. mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2018).
The SAM/BAM Format Specification Working Group Sequence Alignment/Map Format Specification (GitHub, 2021); http://samtools.github.io/hts-specs/SAMv1.pdf
Wang, Z., Weissman, T. & Milenkovic, O. smallWig: parallel compression of RNA-seq WIG files. Bioinformatics 32, btv561 (2015).
Li, H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851 (2014).
ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Hou, H., Quinlan, A. & Pedersen, B. Efficient analysis of quantitative genomics data with the D4 format. Zenodo https://doi.org/10.5281/ZENODO.4684595 (2021).
Acknowledgements
We acknowledge helpful comments from members of the Quinlan laboratory, as well as funding from the National Institutes of Health (NIH) grants HG006693, HG009141 and GM124355 awarded to A.Q.
Author information
Authors and Affiliations
Contributions
H.H. conceived of the D4 encoding strategy, proposed the associated algorithms, implemented all software, conducted all analyses and contributed to the manuscript. B.P. described the original problem that led to the D4 format and contributed to the manuscript. A.Q. supervised the project and led the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Computational Science thanks Christopher Lee, Mikel Hernaez, Christoph Lange and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Handling editor: Fernando Chirigati, in collaboration with the Nature Computational Science team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2, Table 1 and notes.
Source data
Source Data Fig. 1
Depth histogram data for Fig. 1.
Source Data Fig. 3
Primary table sizes as a function of the choice of k for Fig. 3.
Source Data Fig. 4
Times required to create and query files for Fig. 4.
Source Data Fig. 5
Sizes and analysis times from multiple datasets for Fig. 5.
Rights and permissions
About this article
Cite this article
Hou, H., Pedersen, B. & Quinlan, A. Balancing efficient analysis and storage of quantitative genomics data with the D4 format and d4tools. Nat Comput Sci 1, 441–447 (2021). https://doi.org/10.1038/s43588-021-00085-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-021-00085-0
This article is cited by
-
Towards scalable genomic data access
Nature Computational Science (2021)