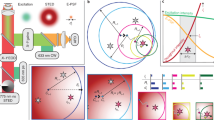
Abstract
Super-resolution fluorescence microscopy is a widely used technique in cell biology. Stimulated emission depletion (STED) microscopy enables the recording of multiple-color images with subdiffraction resolution. The enhanced resolution leads to new challenges regarding colocalization analysis of macromolecule distributions. We demonstrate that well-established methods for the analysis of colocalization in diffraction-limited datasets and for coordinate-stochastic nanoscopy are not equally well suited for the analysis of high-resolution STED images. We propose optimal transport colocalization, which measures the minimal transporting cost below a given spatial scale to match two protein intensity distributions. Its validity on simulated data as well as on dual-color STED recordings of yeast and mammalian cells is demonstrated. We also extend the optimal transport colocalization methodology to coordinate-stochastic nanoscopy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data57 used to create the figures in the main text as well as in the supplement can be found in the Zenodo archive at https://doi.org/10.5281/zenodo.4553856 as well as in the GitHub repository. The data for all figures and Extended Data figures are available in Source Data.
Code availability
The code58 is available on GitHub. The specific version of the OTC package and the scripts generating all figures in this paper can be found at https://doi.org/10.5281/zenodo.4553632. To speed up computation we used the solver CPLEX (v12.6.3.0)59. This IBM product is free for academic use. To download the solver sign up for the IBM academic initiative and download the solver afterwards. To use the solver, download the transport package50 from CRAN as a tar.gz file and change the settings in the makevars file before installing the package. To reproduce any results from the paper please just run the respective script. Without the CPLEX solver, the runtime may take much longer or will not terminate on a standard laptop. With the CPLEX solver, the script for Fig. 5 requires less than 10 min runtime on a standard laptop. If you want to use the OTC package with your own data please see the read me on GitHub.
References
Sahl, S. J., Hell, S. W. & Jakobs, S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 18, 685–701 (2017).
Sigal, Y. M., Zhou, R. & Zhuang, X. Visualizing and discovering cellular structures with super-resolution microscopy. Science 361, 880–887 (2018).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Hess, S. T., Girirajan, T. P. K. & Mason, M. D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Hell, S. W. Far-field optical nanoscopy. Science 316, 1153–1158 (2007).
Klar, T. A., Jakobs, S., Dyba, M., Egner, A. & Hell, S. W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl Acad. Sci. USA 97, 8206–8210 (2000).
Hofmann, M., Eggeling, C., Jakobs, S. & Hell, S. W. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc. Natl Acad. Sci. USA 102, 17565–17569 (2005).
Demandolx, D. & Davoust, J. Multicolour analysis and local image correlation in confocal microscopy. J Microsc. 185, 21–36 (1997).
Bolte, S. & Cordelières, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006).
Worz, S. et al. 3D geometry-based quantification of colocalizations in multichannel 3D microscopy images of human soft tissue tumors. IEEE Trans. Med. Imaging 29, 1474–1484 (2010).
Zinchuk, V. & Grossenbacher-Zinchuk, O. Quantitative colocalization analysis of fluorescence microscopy images. Curr. Protoc. Cell Biol. 62, 4.19.1–4.19.14 (2014).
Costes, S. V. et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86, 3993–4003 (2004).
Dunn, K. W., Kamocka, M. M. & McDonald, J. H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300, C723–C742 (2011).
Barlow, A. L., MacLeod, A., Noppen, S., Sanderson, J. & Guérin, C. J. Colocalization analysis in fluorescence micrographs: verification of a more accurate calculation of Pearson’s correlation coefficient. Microsc. Microanal. 16, 710–724 (2010).
Comeau, J. W. D., Costantino, S. & Wiseman, P. W. A guide to accurate fluorescence microscopy colocalization measurements. Biophys. J. 91, 4611–4622 (2006).
Manders, E. M., Stap, J., Brakenhoff, G. J., van Driel, R. & Aten, J. A. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 103, 857–862 (1992).
Manders, E. M. M., Verbeek, F. J. & Aten, J. A. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382 (1993).
Rizk, A. et al. Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat. Protoc. 9, 586–596 (2014).
Wang, S., Fan, J., Pocock, G. & Yuan, M. Structured correlation detection with application to colocalization analysis in dual-channel fluorescence microscopic imaging. Statistica Sinica. 31, 333–360 (2021).
Wang, S. et al. Spatially adaptive colocalization analysis in dual-color fluorescence microscopy. Preprint at http://arxiv.org/abs/1711.00069 (2017).
Coltharp, C., Yang, X. & Xiao, J. Quantitative analysis of single-molecule superresolution images. Curr. Opin. Struct. Biol. 28, 112–121 (2014).
Georgieva, M. et al. Nanometer resolved single-molecule colocalization of nuclear factors by two-color super resolution microscopy imaging. Methods 105, 44–55 (2016).
Lehmann, M. et al. Quantitative multicolor super-resolution microscopy reveals tetherin HIV-1 interaction. PLoS Pathog. 7, e1002456 (2011).
Malkusch, S. et al. Coordinate-based colocalization analysis of single-molecule localization microscopy data. Histochem. Cell Biol. 137, 1–10 (2012).
Lagache, T., Sauvonnet, N., Danglot, L. & Olivo-Marin, J.-C. Statistical analysis of molecule colocalization in bioimaging. Cytometry A 87, 568–579 (2015).
Lagache, T. et al. Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics. Nat. Commun. 9, 698 (2018).
Mukherjee, S., Gonzalez-Gomez, C., Danglot, L., Lagache, T. & Olivo-Marin, J. Generalizing the statistical analysis of objects’ spatial coupling in bioimaging. IEEE Signal Proces. Lett. 27, 1085–1089 (2020).
Ripley, B. D. The second-order analysis of stationary point processes. J. Appl. Probab. 13, 255–266 (1976).
Helmuth, J. A., Paul, G. & Sbalzarini, I. F. Beyond co-localization: inferring spatial interactions between sub-cellular structures from microscopy images. BMC Bioinformatics 11, 372 (2010).
Shivanandan, A., Radenovic, A. & Sbalzarini, I. F. MosaicIA: an ImageJ/Fiji plugin for spatial pattern and interaction analysis. BMC Bioinformatics 14, 349 (2013).
Blom, H. et al. Nearest neighbor analysis of dopamine D1 receptors and Na(+)-K(+)-ATPases in dendritic spines dissected by STED microscopy. Microsc. Res. Tech. 75, 220–228 (2012).
Levet, F. et al. A tessellation-based colocalization analysis approach for single-molecule localization microscopy. Nature Commun. 10, 2379 (2019).
Zaritsky, A. et al. Decoupling global biases and local interactions between cell biological variables. eLife 6, e22323 (2017).
Peyré, G. & Cuturi, M. Computational optimal transport. Preprint at http://arxiv.org/abs/1803.00567 (2018).
Klatt, M., Tameling, C. & Munk, A. Empirical regularized optimal transport: statistical theory and applications. SIAM J. Math. Data Sci. 2, 419–443 (2020).
Monge, G. Mémoire sur la théorie des déblais et des remblais (De l’Imprimerie Royale, 1781).
Kantorovich, L. V. On a problem of Monge. Usp. Mat. Nauk 3, 225–226 (1948).
Villani, C. Optimal Transport: Old and New (Springer Science & Business Media, 2008).
Sommerfeld, M. & Munk, A. Inference for empirical Wasserstein distances on finite spaces. J. R. Stat. Soc. B 80, 219–238 (2018).
Wang, S., Arena, E. T., Eliceiri, K. W. & Yuan, M. Automated and robust quantification of colocalization in dual-color fluorescence microscopy: a nonparametric statistical approach. IEEE Trans. Image Process. 27, 622–636 (2018).
Göttfert, F. et al. Coaligned dual-channel STED nanoscopy and molecular diffusion analysis at 20 nm resolution. Biophys. J. 105, L01–L02 (2013).
Jans, D.C. et al. STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc. Natl Acad. Sci. USA 110, 8936–8941 (2013).
Stoldt, S. et al. Spatial orchestration of mitochondrial translation and OXPHOS complex assembly. Nat. Cell Biol. 20, 528–534 (2018).
Vogel, F., Bornhövd, C., Neupert, W. & Reichert, A. S. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 175, 237–247 (2006).
de Chaumont, F. et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods 9, 690–696 (2012).
Balzarotti, F. et al. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355, 606–612 (2017).
Schmitzer, B. A sparse multiscale algorithm for dense optimal transport. J. Math. Imaging Vis. 56, 238–259 (2016).
R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2018); https://www.r-project.org/
Schuhmacher, D. et al. Transport: optimal transport in various forms. R package version 0.9-4 https://CRAN.R-project.org/package=transport (2017).
Billingsley, P. Convergence of Probability Measures (Wiley, 2013).
Harke, B. et al. Resolution scaling in STED microscopy. Opt. Express 16, 4154–4162 (2008).
Lagache, T. Colocalization Studio in ICY. ICY http://icy.bioimageanalysis.org/plugin/colocalization-studio/ (2021).
Kehrein, K. et al. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 10, 843–853 (2015).
Melin, J. et al. Presequence recognition by the Tom40 channel contributes to precursor translocation into the mitochondrial matrix. Mol. Cell. Biol. 34, 3473–3485 (2014).
Wurm, C. A., Neumann, D., Schmidt, R., Egner, A. & Jakobs, S. in Live Cell Imaging: Methods in Molecular Biology (ed. Papkovsky, D.) 185–199 (Humana Press, 2010).
Tameling, C. et al. Simluated and real data. Zenodo https://doi.org/10.5281/zenodo.4553856 (2021).
Tameling, C. & Naas, J. ctameling/OTC: optimal transport colocalization. Zenodo https://doi.org/10.5281/zenodo.4553632 (2021).
IBM ILOG CPLEX Optimization Studio (IBM, 2018); https://www.ibm.com/de-de/marketplace/ibm-ilog-cplex
Acknowledgements
We thank P. Rehling from the University Medical Center Göttingen for providing an antiserum specific to Tom40. We are grateful to R. Schmitz-Salue for excellent technical assistance. Further thanks to J. Keller-Findeisen and F. Werner for helpful discussions about the simulation of STED images and to B. Schmitzer on the shielding algorithm. C.T. and J.N. gratefully acknowledge support by the DFG RTN 2088 Project A1. S.J. and A.M. acknowledge support of the DFG Cluster of Excellence MBExC 2067 and DFG-CRC 1456, Project C06. S.J. acknowledges support from the European Research Council (ERCAdG No. 835102).
Author information
Authors and Affiliations
Contributions
A.M. and C.T. developed statistical methodology and algorithms. Furthermore, they performed computer experiments and art work jointly with J.N. S.J., T.S. and S.S. performed experiments and analyzed data jointly with A.M. and C.T. S.J., A.M. and C.T. wrote the manuscript with contributions from all authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Computational Science thanks Thomas Huser, Suvadip Mukherjee, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Ananya Rastogi was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–16, Tables 1–20 and additional simulations.
Supplementary Video 1
Simulation of increasing resolution and influence on Manders’ method and Pearson’s method.
Supplementary Video 2
Visualization of optimal transport between images Tom40 and Mprl4 in yeast cells.
Supplementary Video 3
Visualization of optimal transport of images of two different stainings of Tom40 in yeast cells.
Source data
Source Data Fig. 4
Statistical and image source data.
Source Data Fig. 5
Statistical and image source data.
Source Data Fig. 6
Statistical and image source data.
Source Data Fig. 7
Statistical and image source data.
Rights and permissions
About this article
Cite this article
Tameling, C., Stoldt, S., Stephan, T. et al. Colocalization for super-resolution microscopy via optimal transport. Nat Comput Sci 1, 199–211 (2021). https://doi.org/10.1038/s43588-021-00050-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-021-00050-x
This article is cited by
-
Activation of multiple Eph receptors on neuronal membranes correlates with the onset of optic neuropathy
Eye and Vision (2023)
-
Kantorovich–Rubinstein Distance and Barycenter for Finitely Supported Measures: Foundations and Algorithms
Applied Mathematics & Optimization (2023)
-
Limit laws for empirical optimal solutions in random linear programs
Annals of Operations Research (2022)