Abstract
Traces of heavy metals found in water resources, due to mining activities and e-waste discharge, pose a global threat. Conventional treatment processes fail to remove toxic heavy metals, such as lead, from drinking water in a resource-efficient manner when their initial concentrations are low. Here, we show that by using the yeast Saccharomyces cerevisiae we can effectively remove trace lead from water via a rapid mass transfer process, called biosorption, achieving an uptake of up to 12 mg lead per gram of biomass in solutions with initial lead concentrations below 1 part per million. Through spectroscopic analyses, we found that the yeast cell wall plays a crucial role in this process, with its mannoproteins and β-glucans being the key potential lead adsorbents. Furthermore, by employing nanomechanical characterization in the yeast biomass, we discovered that biosorption is linked to an increase in cell wall stiffness. These findings open new opportunities for using environmentally friendly and abundant biomaterials for advanced water treatment targeting emerging contaminants.
Similar content being viewed by others
Introduction
Heavy metals are highly water-soluble and non-biodegradable, tending to persist indefinitely when released into water bodies. Electronic waste (e-waste) discharge and mining are the most dominant anthropogenic activities responsible for heavy metal contamination of water resources1. Acid mine drainage (AMD), i.e., leakage of highly acidic water rich in metals, is a global environmental threat2. In the United States (US) alone, AMD is the main source of water pollution, impacting currently over 20,000 km of streams3, deriving from the 13,000 active and the 500,000 abandoned mines4, which continue generating AMD for centuries after their closure5.
Lead (Pb) is one of the most widely used heavy metals; its production increased by about 20% during the last decade, reaching around 11.7 million tons globally in 20206. Pb is highly toxic, even at trace concentrations, with deleterious effects on organs and tissues of the human body7. It can enter drinking water either due to inadequate water treatment, or due to chemical reactions with Pb-containing components of water distribution systems8,9. After numerous incidents of Pb contamination8,10, such as the water crisis in the city of Flint, Michigan, US in 2014, limits of Pb in drinking water are becoming more stringent: in 2020 the European Union halved Pb limits in drinking water to 5 parts per billion (ppb)11, while the US Environmental Protection Agency determined that no level of Pb in drinking water is safe12.
Mean Pb concentrations in global surface water bodies are more than 10 times greater than they were in the 1970s, ranging from around 10 ppb in Europe to hundreds of ppb in South America1. Conventional water treatment methods either fail to completely remove trace Pb amounts or result in high financial and environmental costs to do so2,13,14,15. Biosorption, a mass transfer process by which an ion or molecule binds onto inactive biological materials by physicochemical interactions, is a competitive alternative to conventional processes for heavy metal removal, as abundant biomass sources can be effective, practical, and sustainable adsorbents16. Biosorption does not include metabolically mediated sequestering processes of ions or molecules, which are described as bioaccumulation and bioprecipitation16.
Biosorption of heavy metals, and of Pb in particular, has been studied at the parts per million (ppm) contaminant concentration for decades, using different microbial species as candidate biosorbents16 (an overview of prior studies on Pb biosorption at the ppm scale is provided in Supplementary Table 1). However, the mechanisms of metal biosorption are complicated and still not fully understood17, with only a small subset of the work reported in the literature devoted to their systematic and explicit elucidation16. The initial metal ion concentration has a strong effect on the biosorption capacity, while decreasing initial concentrations are linked with decreasing uptake capacities from the biosorbents, if the amount of microbial biomass is kept unchanged17. This fact combined with the absence of information on Pb biosorption isotherm, kinetics and mechanisms at the ppb contaminant concentration, indicate that there is a research gap at trace initial concentrations of ppb and below, and it is worth researching if biosorption can be an efficient process at such initial concentration levels.
In this study, the unexplored Pb biosorption mechanisms at the ppb scale are investigated, using inactive yeast biomass as the biosorbent. A strain of the common yeast, Saccharomyces cerevisiae (S. cerevisiae), was selected as the ideal biosorbent for this investigation, due to its unique nature compared to other microorganisms and fungal species17. S. cerevisiae is a non-pathogenic microorganism, which can be easily cultivated at large scales or obtained in large quantities as a waste or byproduct of various industrial settings14,17,18. It can effectively remove Pb at ppm initial concentrations, and the fact that it has been fully studied as a model system in molecular biology facilitates the investigation of molecular mechanisms involved in biosorption17.
The yeast cells were harvested at the peak of their exponential growth phase (Fig. 1a), for optimal biosorptive capacity17. The harvested cells were washed to remove culture medium residues and metabolites (Fig. 1b), before being lyophilized (freeze-dried) and converted to powder (Fig. 1c). Kinetic and equilibrium experiments were conducted by adding this yeast powder biomaterial to ultrapure water spiked with lead(II) nitrate (Pb(NO3)2). After the required contact time, liquid and solid phases were separated and analyzed to measure residual Pb concentrations and identify potential biomass sites responsible for Pb uptake (Fig. 1d). Overall, this work showcases the use of an effective trace heavy metal removal biomaterial, made from an environmentally friendly, inexpensive, benign to human health, and easy-to-mass-produce microorganism.
a Measured growth curve of S. cerevisiae yeast cells (absorbance accuracy is approximately 1.75% of reported values). b High-performance liquid chromatography (HPLC) analysis results, performed to identify the number of washes with ultrapure water required to remove medium residues and metabolites from harvested yeast cells before being used in biosorption experiments: i) pure culture medium showed a peak at 8 mins representing glucose; ii) in the supernatant of the harvested liquid culture, glucose was not present but ethanol was produced (peak at 18 mins); iii) in the supernatant after the first wash of yeast cells with ultrapure water there were almost no peaks representing the presence of sugars or organic acids in the solution; iv) in the supernatant after the second wash of yeast cells with ultrapure water there were no peaks at all. Therefore, it was decided to wash the harvested cells twice to ensure absence of organic compounds that might affect further experiments. c Yeast powder after lyophilization and scanning electron microscopy (SEM) imaging of freeze-dried yeast cells (scale bar: 5 μm). d Main steps of kinetic and equilibrium experiments involving the addition of freeze-dried yeast cells in Pb-containing aqueous solutions, the adsorption of Pb ions, and the separation of biomass and supernatant after the required contact time via centrifugation for further analyses.
Results
Effect of solution pH on Pb speciation and uptake
Solution pH is a key parameter for biosorption, as it affects the chemistry and speciation of both the metal-uptaking functional groups in the biomass, and of the hydrolyzed Pb ionic forms. Pb speciation in the solution is also affected by the Pb concentration at any given pH and oxidation state16,17,19. To quantify the resulting Pb speciation after the hydrolysis of Pb(NO3)2 in a wide pH range (3–13), at 25 oC, and at a given initial concentration (C0) of 1 μM Pb(NO3)2, the Eawag ChemEQL v3.2 software20 was used. Pb2+ is the dominant species until pH reaches 5.8, where lead hydroxides (e.g., PbOH+) are beginning to form (Fig. 2a).
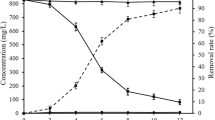
a Distribution of Pb(NO3)2 hydrolysis products at 25 oC and 1 μM Pb(NO3)2. b Increase in solution pH due to yeast biomass addition; error: ± 0.06 pH units. c Effect of initial pH of solution on Pb2+ uptake for 5 mg of yeast biomass and C0 of 100 ppb Pb2+. The final pH of solutions after biosorption is reported for each case; error: ± 0.06 pH units. d Kinetic experiments of 5 mg yeast biomass with C0 of 100 ppb Pb2+, indicating the rapid biosorption process; qt: Pb2+ uptake capacity of yeast biomass at different time intervals (μg of Pb2+ by g of biomass). Time course micrographs of lyophilized S. cerevisiae cells incubated in the same aqueous solution at 0 h and 24 h are shown in the inset, validating that there is no cell growth or division during the experiments (scale bar: 8 μm). e Adsorption isotherm at 25 oC, following the Langmuir adsorption isotherm model; qe: Pb2+ uptake capacity of yeast biomass at equilibrium (μg of Pb2+ by g of biomass). f Pb2+ percentage removal versus Pb2+ C0.
Solution pH increases with the addition of biomass and plateaus for biomass values greater than 100 mg (Fig. 2b). Therefore, to assess the effect of solution pH on the biomass Pb2+ uptake capacity (q, in μg of Pb2+ by g of biomass), the initial pH of the aqueous solution (before biomass addition) was adjusted to pH values within the range of 3–7, and then 5 mg of yeast biomass were added to moderate the anticipated pH increase due to biomass addition. Pb2+ concentrations and pH values were measured both before biomass addition and after Pb2+ biosorption (contact time of 24 h). The biomass Pb2+ uptake capacity greatly increased as the initial solution’s pH was increased from 3 to 5 (Fig. 2c). For pH ≥ 6.0 the measured initial Pb2+ concentration was notably lower than the known amount added to the solutions, indicating loss of soluble Pb analytes due to precipitation. This is validated by the formation of lead hydroxides after pH 5.8 (Fig. 2a). The increase of solution pH by biomass addition, as well as the increase of q with increasing pH values could be attributed to a potential protonation of the functional groups of yeast biomass at pH values below the pH point of zero charges, i.e., the pH at which the overall biomass surface charge is zero. At pH values below the pH point zero, the biomaterial may exhibit an overall positive charge, thus attracting negatively charged species and not adsorbing Pb2+cations resulting in lower q values, while at higher pH values the biomass surface could acquire negative charges leading to increased Pb2+ uptake16. Indeed, the point of zero charges for S. cerevisiae cells is reported in the literature21,22 to be around pH 4.0, hence, the surface of the yeast cells is possible to carry negative charges after this pH value, attracting Pb2+ cations. However, such an approach assumes a rather simplistic electrostatic attraction driving mechanism, which has been shown not to be the only case in biosorption16.
Based on these results, pH 5 was proven to be the most suitable for Pb biosorption, where soluble Pb2+ is the most dominant species in the solution and q is maximized. The pH region around 5 has also been proven to be the most suitable for optimal uptake capacities for Pb concentrations at the ppm scale17. All experiments described in the following sections were performed by adding 5 mg of yeast biomass (unless otherwise specified) in aqueous solutions, after adjusting the initial solution pH to 5. Solutions were then incubated at 25 oC and agitated at 200 rpm.
Adsorption kinetics & growth analysis of lyophilized yeast
Kinetic experiments were conducted to determine the change in Pb2+ concentration in the liquid phase as a function of contact time and identify the contact time required to attain equilibrium. Lyophilized yeast cells were added in aqueous solutions with C0 of 100 ppb Pb2+ and were incubated for 24 h. Samples were taken at specific time intervals (i.e., 0 min, 5 min, 15 min, 30 min, 1 h, 2 h, 4 h, 8 h, 24 h) and analyzed using inductively coupled plasma mass spectrometry (ICP-MS). As observed, biosorption is a rapid process, with equilibrium being reached within the first five minutes of contact (Fig. 2d).
The widely applied pseudo-first-order23 and the pseudo-second-order24 kinetic models were assessed for fitting the experimental data. Both the pseudo-first-order and the pseudo-second-order model fitted well the experimental kinetic data of the present study, but the pseudo-first-order model provided a slightly better description of the measured values (R2: 0.99, all other statistical parameters closer to 0). The high value of the pseudo-first-order rate constant (k1), which is equal to 111.98 h−1, confirms the rapid achievement of the adsorption equilibrium. More information on the fitting of adsorption kinetic models and relevant parameters is provided in the Supplementary Information (Supplementary Note 1, Supplementary Fig. 1, and Supplementary Table 2). However, these models are empirical, and they do not have any mechanistic substrate16 so as to elucidate the biosorption mechanisms. The only conclusion that can be drawn by the kinetic parameters is that Pb2+ biosorption at the ppb scale is a rapid phenomenon, which is highly advantageous for engineering applications.
In parallel, the freeze-dried S. cerevisiae cells were incubated for 24 h under conditions identical to the kinetic experiments and observed by phase-contrast microscopy, while acquiring optical density (OD) measurements, to validate that the cells remain inactive during biosorption. Indeed, after 24 h there was neither cell growth nor cell division observed (Fig. 2d, Supplementary Fig. 3, Supplementary Movies 1 and 2).
Adsorption isotherm
Aqueous solutions with different initial Pb2+ concentrations (C0: 20, 40, 100, 200, 300, 500, 700, and 1000 ppb) were incubated for 1 h. The equilibrium Pb2+ concentrations (Ceq) were measured after biosorption via ICP-MS, and the Pb2+ uptake capacity of yeast biomass at equilibrium (qe) was calculated (using Eq. (1) in Methods section) to develop the adsorption isotherm (Fig. 2e). The maximum qe measured is about 12 mg/g, for aqueous solutions with C0 of 1000 ppb Pb2+.
The commonly used isotherm models of Langmuir25 and Freundlich26, borrowed from the ideal adsorption theory of gases, were assessed for fitting the experimental data. The experimental data are accurately fitting the Langmuir adsorption isotherm model (R2: 0.98), with the ratio of the adsorption and desorption rates (KL) equal to 1.5 L/mg, and the maximum estimated adsorption capacity (qm) equal to 22.5 mg/g. More information on the fitting of adsorption isotherm models and relevant parameters is provided in the Supplementary Information (Supplementary Note 2, Supplementary Fig. 2). However, in contrast to the ideal adsorption theory of gases, these models bear no mechanistic significance for biosorption, and this fit cannot provide any meaningful insight regarding the underlying mechanisms16,17.
Pb2+ percentage removal versus Pb2+ C0 was also measured (Fig. 2f). Pb2+ removal for C0 of 20 ppb Pb2+ is approximately 25% and increases with the increase of C0, reaching a maximum of 43% at C0 of 300 ppb Pb2+. After this point, Pb2+ removal decreases gradually with the increase of C0, indicating that the optimum uptake capacity of the yeast biomass quantity (5 mg) is reached around C0 of 300 ppb Pb2+.
Yeast biomass imaging
Extracellular and intracellular imaging of yeast cells was performed to observe potential changes in their structure after biosorption, using scanning electron microscopy (SEM) imaging and transmission electron microscopy (TEM) imaging, respectively. Yeast cells harvested from ultrapure water (C0: 0 ppb Pb2+), served as control cells and were compared with yeast cells harvested from aqueous solutions of C0 100 ppb Pb2+. Given a q of ~1250 ug/g at this C0, based on the kinetics and adsorption isotherm experiments, Pb2+ concentration in the yeast cells harvested from aqueous solutions with C0 of 100 ppb Pb2+ was about 0.12% wt. No morphological change was observed in the yeast cells after Pb2+ biosorption, with the structure and dimensions of the cell wall and cytoplasm remaining the same (Fig. 3a, b). Yeast cell walls were ~180 nm thick, which is the typical cell wall thickness of the yeast strain used27. As Pb2+ concentration was very close to the TEM detection limit (0.1% wt.), Pb2+ deposition on the yeast cells could not be clearly visualized in contrast with studies of initial Pb2+ concentrations at the ppm scale (Pb2+ concentration in biomass of 1–2 orders of magnitude higher than the present study)28,29. Therefore, no valuable conclusion regarding Pb2+ uptake could be drawn from the TEM images at such low concentration levels (Fig. 3c).
Rows are ordered as follows: control yeast cells (C0: 0 ppb) (1), yeast cells after Pb2+ biosorption (C0: 100 ppb) (2). a SEM images presenting an overview of yeast cells (scale bar: 10 μm). b Magnified SEM images showing individual yeast cells (scale bar: 2 μm). c TEM images of individual yeast cells (scale bar: 1 μm).
Yeast biomass spectroscopy
Metal biosorption is thought to occur through interactions with functional groups native to the biomass cell wall30. Attenuated total reflectance Fourier transform infrared spectroscopy (ATR-FTIR) was performed to identify functional groups present in the yeast cell wall and detect changes in them after biosorption, indicating their involvement in Pb2+ adsorption. Freeze-dried control (C0: 0 ppb Pb2+) and Pb2+-exposed yeast cells (C0: 100 and 1000 ppb Pb2+) were analyzed using ATR-FTIR (Fig. 4a). Changes were observed after biosorption in peaks representing C ≡ N and C ≡ C stretches, while peak shifts were detected corresponding to N-H in-plane bending from secondary protein amides, which overlaps with the C–N and NO2 asymmetric stretching, to vibrational changes of the C-N amide group, and to C–O stretching in the esters and carboxylic acid groups. These changes indicate the contribution of amide and carboxylic acid groups to Pb2+ biosorption and the potential role of N in the yeast cell wall in Pb2+ binding. Detailed description of the ATR-FTIR results is provided in Supplementary Note 3. All observed changes in ATR-FTIR peaks before and after Pb2+ biosorption are shown in Supplementary Table 3 and Supplementary Fig. 5.
a Full ATR-FTIR spectrum of yeast cells. The black line represents control yeast cells (C0:0 ppb Pb2+); the blue and green lines represent yeast cells after biosorption with C0 of 100 and 1000 ppb Pb2+ respectively. b XPS analysis of yeast cells. From left to right: control cells, yeast cells after biosorption, C0 100 and 1000 ppb Pb2+. XPS peak assignments are as follows: light green C=O; magenta: O-C=O; purple: C-N; dark green: C-C.
In addition, the chemical composition of the yeast surface before (C0: 0 ppb Pb2+) and after biosorption (C0: 100 and 1000 ppb Pb2+) was analyzed by X-ray photoelectron spectroscopy (XPS) to further explore potential changes in the functional groups of the yeast cell walls. The yeast surface is mainly composed of carbon (C), oxygen (O), and nitrogen (N)31,32 (Supplementary Table 4). Therefore, C 1 s, O 1 s, N 1 s core levels spectra were recorded together with the Pb 4 f. Changes among the control and the Pb2+-exposed yeast were only observed for the C 1 s spectrum, while changes in the Pb spectra could not be detected as the analyzed trace Pb2+ concentrations were below the detection limit of the instrument (Supplementary Table 5). Deconvolution of the C 1 s spectrum into Gaussian-shaped lines was performed to identify possible chemical bonds between C, O, and N (Fig. 4b). In all three samples, the C 1 s peaks are decomposed to peaks at 284.3 eV, 285.9 eV, 287.5 eV, and 288.6 eV representing C-C (sp3 C), C-N, O-C=O, and C=O bonds respectively. The magnitude and shape of all observed bonds have radically changed after Pb2+ biosorption, particularly for C-C, C-N, and O-C=O bonds. These changes indicate the contribution of carboxylic acid and amide groups to Pb2+ adsorption, which is consistent with the ATR-FTIR results. Changes in carboxyl, amino, hydroxyl and amide groups of protein and carbohydrate fractions of yeast cell walls have been also reported in the literature for higher initial Pb2+ concentrations (ppm scale)17,33.
Chitin’s contribution to Pb2+ adsorption
The above analyses indicate that the cell wall of S. cerevisiae plays a vital role in Pb2+ biosorption. The yeast cell wall has a complex macromolecular structure with a layered organization, including an amorphous inner and a fibrillar outer layer33. The inner layer mainly consists of β-glucans and chitin. The outer layer consists predominantly of mannan polymers, highly glycosylated and linked to proteins (mannoproteins)34 (Supplementary Fig. 6).
Several sources suggest that the chitin amine nitrogen is responsible for heavy metals sequestering at the ppm scale16,35,36. To further investigate this, we assessed the Pb2+ uptake capacity of chitin from shrimp shells, which has similar characteristics to the chitin found in the yeast cell wall37,38,39. We used 20 times more chitin than the maximum equivalent amount present in the 5 mg of yeast, i.e., 1.8 mg, considering a 30% dry weight of yeast cell wall and a 6% contribution of chitin per mass to it34. We added this amount to an aqueous solution of 0.2 L with C0 of 1000 ppb Pb2+ for 24 h at 200 rpm and 25 oC. We ran the same experiments with 5 mg of yeast biomass and with 5 mg of chitin. It was shown that chitin’s Pb2+ uptake is negligible, as C0 was reduced by less than 0.3% when we added the 1.8 mg of chitin, which is within the measurement error. Even when the chitin amount added was equal to the total yeast mass (5 mg), C0 was only reduced by 3%, compared to the ~30% reduction achieved by the yeast biomass (Fig. 5a). Hence, it can be concluded that chitin alone is not contributing to the Pb2+ biosorption process.
a Pb2+ Ce after biosorption experiments with 5 mg of yeast biomass (green bar), 1.8 mg, and 5 mg of chitin (purple bars); blue dotted line represents C0: 1000 ppb Pb2+. b AFM analysis of yeast cell stiffness. Box and whisker plots of the cellular spring constant (kcell) calculated from force-extension curves acquired from yeast cells incubated with C0 of 0, 500, and 1000 ppb Pb2+. Boxed region indicates upper and lower quartiles for each data set; median is indicated by the horizontal line within the box; mean is indicated by the bullet point; whiskers extend to high and low data points (n > 70 force measurements from ≥ 8 cells per condition). Asterisk indicate p < 1e-5 by single factor ANOVA test between cell groups incubated with C0 of 0 ppb and 500 ppb, or 1000 ppb Pb2+. AFM deflection retrace image of yeast cells is shown in the inset (scale bar: 2 μm).
Yeast biomass nanomechanical characterization
We employed nanomechanical characterization to investigate biosorption. We assessed the stiffness of the yeast cells before and after Pb2+ exposure, by conducting shallow (<10% of hydrated cell wall thickness) atomic force microscopy (AFM) indentations. Single-cell AFM mechanical testing showed a notable increase in the stiffness of cells following Pb2+ uptake (Fig. 5b, Supplementary Fig. 7). As shallow AFM indentations provide an isolated cell wall response rather than a bulk cell response, AFM results confirm Pb2+ ion reinforcement of the yeast cell wall. It is possible that adsorption of a thin layer of Pb2+ can act as a film on the cell surface that fuses the fibrillar structures together, which then effectively resists deformation more than the untreated cell wall. However, when yeast biomass is treated with solutions containing higher Pb2+ levels, mechanical stiffness is not noticeably increased (Fig. 5b, C0 of 500 vs 1000 ppb Pb2+). This cannot be attributed to a potential saturation of the yeast cell wall binding sites, as Pb2+ uptake increases substantially with the increase of C0 from 500 to 1000 ppb Pb2+, not reaching saturation (Fig. 2e). On the other hand, maximum Pb2+ removal is observed for C0 of 300 ppb, and the Pb2+ uptake rate drops with increasing C0 after this point, indicating lower adsorption efficiency (Fig. 2f). While the exact mechanism for this stiffness change is yet to be determined, from the AFM experiments it can be concluded that the rate of yeast cell wall stiffening follows a similar pattern to that of the Pb2+ uptake rate with increasing C0 (lower rates for C0 > 300 ppb).
Discussion and conclusions
This work reports for the first time, to the best of our knowledge, the biosorption isotherm and kinetics of initial Pb2+ concentration at the ppb scale, using lyophilized S. cerevisiae yeast cells as biosorbents. By comparing our results with prior studies of similar systems at the ppm scale it can be concluded that the biosorption processes at the ppb scale happen faster; the fastest equilibrium attainment reported at the ppm scale is 10 min17, while our results show that equilibrium is achieved within the first 5 minutes of contact. The adsorption isotherm reported in our study (Fig. 2e) follows the same pattern as the adsorption isotherms reported in the ppm literature (Supplementary Fig. 4). Interestingly, the maximum Pb2+ uptake capacity of 12 mg/g reported in this study is in the same range as the uptake capacities reported in the ppm literature for untreated inactive S. cerevisiae yeast, i.e., 2–30 mg/g (Supplementary Table 1), proving the suitability of this biomaterial as a biosorbent at the ppb scale. pH greatly affects the biosorption process in both scales.
The rapid biosorption and high Pb2+ uptake are advantageous for the large-scale application of this inexpensive and abundant biomaterial for the removal of trace heavy metals from water. It is worth investigating its implementation to other heavy metal cations as well, such as copper (Cu2+) and cadmium (Cd2+). However, S. cerevisiae has been proven to have higher affinity with Pb2+ compared to other heavy metals17,18,30, hence, it is expected that the adsorption capacity of other heavy metal cations at the ppb scale would be lower compared to Pb2+. The investigation of potential co-ion effects would be also valuable for the large-scale application of this approach, as various compounds are usually concurrently present in contaminated water. A potential reduction of the Pb2+ adsorption capacity is expected in such case, based on the trends reported in prior studies of similar systems at the ppm scale17,30. However, specific experiments need to be conducted to validate how the biosorption process would be affected by ionic competition, as chemistry and speciation can differ for different metal ions.
From the performed analyses, it can be concluded that a one-step Pb2+ uptake process occurs, mainly due to the cell wall of S. cerevisiae, and in particular its carboxylic acid and amide groups. The cell wall of S. cerevisiae, and particularly its amino, carboxyl, and hydroxyl groups has been shown to greatly contribute to the biosorption of heavy metal cations at the ppm scale as well17,40. Although further elucidation of the exact uptake processes is needed, there is no indication of a multi-step uptake process, which is often reported in the ppm literature for non-living biomass, involving diffusion and/or accumulation of Pb2+ ions inside the cytoplasm17,28,29. As the yeast cells are inactive, additional steps would require disruption of the yeast cell membrane, or changes in its permeability, allowing the penetration of Pb2+ ions. However, freeze-drying of yeast cells protects their structure and is not associated with loss of membrane integrity, in contrast with autoclaving28,29, hence, the occurrence of additional uptake steps is unlikely.
Our chitin experiment provides direct evidence of the importance of proteins and protein-carbohydrates of the outer and middle layers of the cell wall over the chitinous inner layers in Pb2+ biosorption. By excluding chitin as a biosorbent, mannoproteins and β-glucans are the potential key S. cerevisiae cell wall components, which should be further analyzed to elucidate the biosorption mechanisms involved. The contribution of the outer mannan-protein layer of the yeast cell wall in accumulating heavy metal cations, e.g., Cu2+ and Cd2+, has been previously demonstrated40 validating our conclusions.
The combined outcomes of the spectroscopic analyses, and the cellular nanomechanical characterization, validate the likelihood of N-linked σ-hole attraction to Pb2+ species as a possible mechanism of biosorptive Pb2+ retention by the mannoprotein/β-glucan cell wall fraction41,42, leading to supramolecular assemblies that make yeast cells stiffer after biosorption (potential Pb2+ coordination schemes with amide groups have been suggested in the literature42,43). These findings open new experimental pathways for approaching the challenging task of biosorption investigation at the ppb scale.
Exploiting yeast as a biosorbent can be practically feasible and economically attractive. S. cerevisiae can be easily cultivated in large quantities, while having various beneficial industrial applications, e.g., in the food, beverage, therapeutics and biofuel production industries44. Three million tons of yeast are used annually by the global fermentation industry45, while the yeast market is expected to grow by 35% in the next 5 years46. However, the sugars and proteins needed for yeast growth are derived from food crops and sources that could be used for human nutrition. Therefore, to avoid compromising food security, residual or surplus yeast from fermentation industries would be a more sustainable option for the large-scale application of this water purification approach. Indeed, surplus yeast is currently produced in huge volumes and is an extremely underutilized low-value resource, not suitable as a human dietary supplement due to high levels of nucleic acids47.
The approach described here compares favorably to many of the highly sophisticated synthetic biology and advanced nanomaterials approaches that have also been examined as candidates for heavy metal removal from water48,49. Applying such a low-value resource to remove trace contaminants from water could also result in waste reduction as yeast cells are biodegradable. Moreover, potential desorption processes would allow for heavy metals reclamation, enhancing the application of circular economy models.
Methods
Glassware cleaning
Glassware was autoclaved for 15 minutes at 121 °C (tabletop autoclave 3870E, Tuttnauer, USA) and rinsed three times with Type I ultrapure water, with a resistivity of 18.2 MΩ.cm at 25 °C and total organic carbon (TOC) < 5 ppb (Milli-Q Direct 8 Water Purification System, Millipore Sigma, USA). Phosphate-free detergent, suitable for trace heavy metals analyses, was used to wash the glassware (Liquinox detergent, Alconox, USA), which was then rinsed 3 times with ultrapure water and soaked in a 20% nitric acid (HNO3) bath for at least 24 h. The HNO3 bath was made using 69% HNO3 (ARISTAR PLUS, VWR Chemicals BDH, VWR, USA) and ultrapure water. After soaking in the 20% HNO3 bath, flasks were rinsed 3 times with ultrapure water prior to use.
Yeast strain, culture & OD600 culture density measurements
The S. cerevisiae Meyen ex E.C. Hansen MYA-796 strain was used (ATCC, Virginia, USA). YM agar and broth media (ATCC 200 YM Medium, ATCC, Virginia, USA) were used for the yeast cultures. Yeast cells were incubated (Multitron Standard incubator shaker, INFORS HT, USA) at 30 oC and 200 rpm in 2 L Erlenmeyer flasks with shallow medium content (100-200 mL). Discrete time-point OD measurements were performed using 2 mL non-frosted cuvettes (VWR Standard Spectrophotometer Cuvettes, VWR, USA) and a tabletop, ultraviolet-visible spectrophotometer measuring at 600 nm (NanoPhotometer NP80, Implen, Munich, Germany).
Biomass harvesting, washing & lyophilization
Yeast cells were harvested by centrifugation at 1 g/2000 rpm for 10 min (Allegra X15-R Centrifuge, USA) and washed by two successive suspensions and centrifugations with ultrapure water. To decide the number of washes required to remove medium residues and metabolites from cells, supernatant samples after each wash were analyzed using HPLC (Aminex HPX-87H Column, Bio-Rad, USA). Harvested, washed cells were kept at −30 oC for 24 h and then inserted in the freeze dryer (Freezone 6 Liter Manifold, Labconco, USA). Each lyophilization cycle (temperature < −40 oC, pressure < 0.371 mbar) lasted for at least 50 h. Lyophilized cells (powder form) were stored in a desiccator containing silica gel. Freeze-dried biomass was weighted using an analytical balance of 0.1 mg precision and resolution (AB104-S analytical balance, Mettler Toledo, USA).
Aqueous solutions preparation
Type I ultrapure water was spiked with lead(II) nitrate (Pb(NO3)2) (Sigma-Aldrich, Millipore Sigma, USA) to achieve solutions with initial Pb2+ concentrations of up to 1000 ppb. The initial solution pH was adjusted to the required values by using 69% HNO3 (ARISTAR PLUS, VWR Chemicals BDH, VWR, USA) and 0.1 M sodium hydroxide (NaOH) (Carolina Biological, USA). The pH of aqueous solutions was measured using a glass-body electrode suitable for ion-weak samples (InLab Pure Pro-ISM, Mettler Toledo, USA).
Biosorption experiments
All biosorption experiments were conducted in batch contact environments using 2 L Erlenmeyer flasks with lyophilized yeast cells added to 200 mL of Pb2+ containing aqueous solutions. Flasks were incubated at 200 rpm and 25 oC. After the required contact time, yeast biomass was separated from the aqueous solutions by centrifugation at 1 g/2000 rpm for 10 min (Allegra X15-R Centrifuge, USA). The supernatants were analyzed using ICP-MS (7900 ICP-MS system, Agilent, USA) to measure the residual Pb2+ concentrations following standard operating procedures (Pb2+ calibration standards and Bismuth internal standard, Agilent, USA). All experiments were carried out in triplicates and mean values are reported.
For all experiments, a control sample of just ultrapure water spiked with Pb(NO3)2 was measured to act as a reference for the initial Pb2+ concentration, C0, in the solution. Pb2+ removal measurements were calculated by taking the ICP-MS measurements of the supernatants and subtracting from this the reference to determine the quantity of metal adsorbed by yeast biomass. Type I ultrapure water alone was also tested via ICP-MS to make sure that there was no Pb2+ present in the aqueous matrix. The amount of Pb2+ uptake from the yeast biomass was calculated using Eq. (1):
Where: qt is the Pb2+ mass (μg) adsorbed per gram of yeast biomass after t contact time (μg/g); C0 is the initial Pb2+ concentration in the aqueous solution (μg/L or ppb); Ct is the residual Pb2+ concentration measured after t contact time (μg/L or ppb); m is the dry weight of yeast biomass in the solution (g), and V is the volume of the aqueous solution (L). If equilibrium has been reached and Ct is Ce, then qt is qe.
The tools developed by Wang and Guo50,51 were used for solving the nonlinear adsorption kinetic and isotherm models, using the experimental Pb2+ biosorption kinetics data, i.e., the q values measured at specific time intervals (i.e., 0 min, 5 min, 15 min, 30 min, 1 h, 2 h, 4 h, 8 h, 24 h) of contact with yeast biomass (m: 0.005 g) in aqueous solutions (0.2 L) with C0 of 100 ppb, and isotherm data, i.e., the q values at equilibrium measured after 1 h of contact of yeast biomass (m: 0.005 g) with aqueous solutions (0.2 L) of different initial Pb2+ concentrations (C0: 20, 40, 100, 200, 300, 500, 700 and 1000 ppb), respectively.
Biosorption experiments and analyses using chitin were conducted following the same steps, procedures, and instruments described above, using chitin instead of yeast biomass (Chitin from shrimp shells, Sigma-Aldrich, Millipore Sigma, USA).
SEM sample preparation and analysis
After biosorption, yeast biomass was harvested via centrifugation at 1 g/2000 rpm for 10 mins (Allegra X15-R Centrifuge, USA) and lyophilized, following the procedures described above. Samples of lyophilized yeast biomass (before and after Pb2+ biosorption) were coated in a 20 nm layer of gold (150TES Sputter Coater, EMS, USA), mounted onto carbon double-sided tape, and imaged with an SEM, using secondary electron detection at 10 kV beam voltage and varying magnifications (FlexSEM 1000, Hitachi, Tokyo, Japan).
TEM sample preparation and analysis
Harvested yeast cells were fixed with 2% glutaraldehyde and 2.5% paraformaldehyde in 100 mM sodium cacodylate buffer (EMS, USA) for 1 h at 4 oC. Then cells were washed twice with 100 mM sodium cacodylate buffer, each time re-suspending them by vortexing, storing them at 4 oC for 15 mins, and centrifuging them for 10 mins at 1 g/2000 rpm. Then cells were resuspended in 100 mM sodium cacodylate buffer and stayed overnight at 4 oC. Pelleted cells were post-fixed for 30 min at 4 oC with 1% osmium tetroxide in 0.1 M imidazole of pH 7.5 (EMS, USA). Osmium post-fixation was followed by washing the cells three times with 100 mM sodium cacodylate buffer, storing them each time for 15 mins at room temperature. The cells were then washed three times with 0.05 M maleate buffer of pH 5.15 (EMS, USA), storing them each time for 15 mins at room temperature. The cells were then stained in 2% uranyl acetate (EMS, USA) overnight, in the dark at room temperature. Stained cells were washed three times with 0.05 M maleate buffer of pH 5.15, storing them each time for 15 mins at room temperature. Then, the cells were serially dehydrated using 30%, 35%, 50%, 70%, 90%, 95%, and 100% ethanol solutions. Dehydrated cells were embedded in EMBED-812 resin (EMS, USA). Sections were cut using an ultramicrotome (EM UC7, Leica, Wetzlar, Germany) and a Diatome diamond knife at a thickness setting of 50 nm. The sections were examined using a TEM (Tecnai G2 Spirit TWIN, FEI, USA) at 120 kV.
Measurement of yeast cell wall dimensions
Yeast cell walls were measured using Image J software, taking the average measurements of 15 control and 15 Pb2+-exposed cells.
ATR-FTIR analysis
Samples of lyophilized yeast biomass (before and after Pb2+ biosorption) were put on the ATR sampling accessory of an FTIR spectrometer (Nicolet iS50 FTIR Spectrometer, Thermo Fisher Scientific, USA). The measured wavenumber range was 400 − 4000 cm−1, and for all spectra 32 scans were recorded and averaged with a resolution of 4 cm−1 for each spectrum. All the handling material and the ATR sampling surface were cleaned with analytical grade isopropanol and rubbed dry with clean paper before contact with the host material.
XPS analysis
Lyophilized yeast cells were taken with a spatula and loaded on a stainless steel XPS stage (5 mm width). All samples were analyzed in the same XPS run (spot size: 400 μm) (K-Alpha XPS system, Thermo Fisher Scientific, USA). Survey scans used a pass energy of 50 eV, while the core scans used a pass energy of 1.5 keV and were energy-calibrated using the C-C bond energy at 284.6 eV. The three peaks observed in the C 1 s spectrum were fitted to identify changes in the C bonds due to Pb2+ biosorption. Data processing and peak fitting was done using the Thermo Avantage software and the Powel fitting algorithm. All the handling materials and the polyacetal surface were cleaned with analytical grade isopropanol and rubbed dry with clean paper before contact with the host material.
AFM sample preparation and analysis
Lyophilized yeast biomass (following incubation with or without Pb2+ at 200 rpm and 25 °C for 1 h) was harvested via centrifugation at 1 g for 5 mins and adhered to plasma cleaned and CellTak (Corning Inc, USA) adhesive-treated plastic 50 × 9 mm petri dishes, as described elsewhere52. AFM force analysis was conducted in liquid (AFM tests were done in hydrated cells) with an MFP-3D AFM (Asylum Research, USA), using spherical colloidal tip CP-PNP-Au-A probes (NanoAndMore, USA) with a sphere diameter of 2 μm and a nominal spring constant of 0.08 N/m. Cell surface indentation was limited to <10% of the hydrated cell wall thickness with a 1.98 μm/s approach velocity. Cellular spring constants (kcell) were calculated as described elsewhere using a two-spring model52,53.
Figure creation
Raw data were collected and stored in CSV or Excel file formats. Plots were created using the Origin software.
Data availability
The datasets generated and analyzed during the current study are available at https://doi.org/10.6084/m9.figshare.19660698.v454.
References
Zhou, Q. et al. Total concentrations and sources of heavy metal pollution in global river and lake water bodies from 1972 to 2017. Glob. Ecol. Conserv. 22, e00925 (2020).
Klein, R., Tischler, J. S., Mühling, M. & Schlömann, M. Bioremediation of Mine Water. in Geobiotechnology I (eds Schippers, A., Glombitza, F. & Sand, W.) vol. 141, 109–172 (Springer Berlin Heidelberg, 2013).
Acharya, B. S. & Kharel, G. Acid mine drainage from coal mining in the United States – An overview. J. Hydrol. 588, 125061 (2020).
Statista. Number of active mines in the United States from 2000 to 2019. https://www.statista.com/statistics/949127/number-active-mines-united-states-by-commodity/ (2021).
Abandonedmines.gov. Water Pollution. https://www.abandonedmines.gov/water-pollution.
Statista. World production of lead from 2006 to 2020. https://www.statista.com/statistics/264872/world-production-of-lead-metal/ (2021).
Casas, J. S. & Sordo, J. Lead: Chemistry, Analytical Aspects, Environmental Impact and Health Effects. (Elsevier, 2006).
Santucci, R. J. & Scully, J. R. The pervasive threat of lead (Pb) in drinking water: Unmasking and pursuing scientific factors that govern lead release. Proc. Natl. Acad. Sci. 117, 23211–23218 (2020).
Sharma, P. R. et al. Lead removal from water using carboxycellulose nanofibers prepared by nitro-oxidation method. Cellulose 25, 1961–1973 (2018).
WHO. Lead in Drinking-water. Background document for development of WHO Guidelines for Drinking-water Quality. https://www.who.int/water_sanitation_health/dwq/chemicals/lead.pdf (2011).
European Union. Directive (EU) 2020/2184 of the European Parliament and of the Council of 16 December 2020 on the quality of water intended for human consumption. https://eur-lex.europa.eu/eli/dir/2020/2184/oj (2020).
NSF International. Revisions to drinking water standard tighten lead leaching allowance for plumbing products. https://phys.org/news/2020-09-standard-tighten-leaching-plumbing-products.html?utm_source=nwletter&utm_medium=email&utm_campaign=daily-nwletter (2020).
Johnson, D. B. & Hallberg, K. B. Acid mine drainage remediation options: a review. Sci. Total Environ. 338, 3–14 (2005).
Sun, G. L., Reynolds, Erin, E. & Belcher, A. M. Using yeast to sustainably remediate and extract heavy metals from waste waters. Nat. Sustain. 3, 303–311 (2020).
Tian, H., Alkhadra, M. A., Conforti, K. M. & Bazant, M. Z. Continuous and Selective Removal of Lead from Drinking Water by Shock Electrodialysis. ACS EST Water acsestwater.1c00234 (2021) https://doi.org/10.1021/acsestwater.1c00234.
Tsezos, M. Biosorption: A Mechanistic Approach. in Geobiotechnology I: Metal-related Issues (eds Schippers, A., Glombitza, F. & Sand, W.) 173–209 (Springer-Verlag, 2014).
Wang, J. & Chen, C. Biosorption of heavy metals by Saccharomyces cerevisiae: A review. Biotechnol. Adv. 24, 427–451 (2006).
Ojima, Y. et al. Recovering metals from aqueous solutions by biosorption onto phosphorylated dry baker’s yeast. Sci. Rep. 9, 225 (2019).
Baes, C. F. & Mesmer, R. E. The hydrolysis of cations. (Wiley, 1976).
Eawag. ChemEQL - a software for the calculation of chemical equilibria. https://www.eawag.ch/en/department/surf/projects/chemeql/ (2021).
Saitoh, N., Noruma, T. & Konishi, Y. Sustainable use of precious and rare metals through biotechnological recycling. In REWAS 2019: Manufacturing the Circular Materials Economy (The minerals, metals & materials series)(eds G. Gaustad et al.), 107–114, (Springer, Cham., 2019).
Dengis, P. B., Nélissen, L. R. & Rouxhet, P. G. Mechanisms of yeast flocculation: Comparison of top-and bottom-fermenting strains. Appl. Environ. Microbiol. 61, 718–728 (1995).
Lagergren, S. About the theory of so-called adsorption of soluble substances. K. Sven. Vetenskapsakad. Handl. 24, 1–39 (1898).
Ho, Y. S., Wase, D. A. J. & Forster, C. F. Removal of lead ions from aqueous solution using sphagnum moss peat as adsorbent. Water S.A. 22, 219–224 (1996).
Langmuir, I. The constitution and fundamental properties of solids and liquids. Part I. Solids. J. Am. Chem. Soc. 38, 2221–2295 (1916).
Freundlich, H. M. F. Über die adsorption in lösungen. Z. Phys. Chem. 57, 385–470 (1906).
Hudson, L. E. et al. Characterization of the Probiotic Yeast Saccharomyces boulardii in the Healthy Mucosal Immune System. PLOS ONE 11, e0153351 (2016).
El-Sayed, M. T. Removal of lead(II) by Saccharomyces cerevisiae AUMC 3875. Ann. Microbiol. 63, 1459–1470 (2013).
Suh, J. H., Yun, J. W. & Kim, D. S. Comparison of Pb2+ accumulation characteristics between live and dead cells of Saccharomyces cerevisiae and Aureobasidium pullulans. Biotechnol. Lett. 20, 247–251 (1998).
Göksungur, Y., Üren, S. & Güvenç, U. Biosorption of cadmium and lead ions by ethanol treated waste baker’s yeast biomass. Bioresour. Technol. 96, 103–109 (2005).
Dengis, P. B. & Rouxhet, P. G. Preparation of yeast cells for surface analysis by XPS. J. Microbiol. Methods 26, 171–183 (1996).
Dengis, P. B., Genet, M. J. & Rouxhet, P. G. Microbial Cells by XPS: Analysis of Brewing Yeast Saccharomyces cerevisiae. Surf. Sci. Spectra 4, 21–27 (1996).
Machado, M. D., Janssens, S., Soares, H. M. V. M. & Soares, E. V. Removal of heavy metals using a brewer’s yeast strain of Saccharomyces cerevisiae: advantages of using dead biomass. J. Appl. Microbiol. 106, 1792–1804 (2009).
Orlean, P. Architecture and Biosynthesis of the Saccharomyces cerevisiae Cell Wall. Genetics 192, 775–818 (2012).
Tsezos, M. & Mattar, S. A further insight into the mechanism of biosorption of metals, by examining chitin epr spectra. Talanta 33, 225–232 (1986).
Tsezos, M. & Volesky, B. Biosorption of uranium and thorium. Biotechnol. Bioeng. 23, 583–604 (1981).
Di Mario, F., Rapana, P., Tomati, U. & Galli, E. Chitin and chitosan from Basidiomycetes. Int. J. Biol. Macromol. 43, 8–12 (2008).
Afroz, M., Kashem, M., Hasan, N., Piash, K. M. & Islam, N. Saccharomyces cerevisiae as an untapped source of fungal chitosan for antimicrobial action. Appl. Biochem. Biotechnol. 193, 3765–3786 (2021).
Acosta, N., Jiménez, C., Borau, V. & Heras, A. Extraction and characterization of chitin from crustaceans. Biomass Bioenergy 5, 145–153 (1993).
Brady, D., Stoll, A. D., Starke, L. & Duncan, J. R. Chemical and Enzymatic Extraction of Heavy Metal Binding Polymers from Isolated Cell Walls of Saccharomyces cerevisiae. Biotechnol. Bioeng. 44, 297–302 (1994).
Mahmoudi, G. et al. On the importance of tetrel bonding interactions in lead(II) complexes with (iso)nicotinohydrazide based ligands and several anions. Dalton Trans. 45, 10708–10716 (2016).
Politzer, P., Murray, J. S., Clark, T. & Resnati, G. The σ-hole revisited. Phys.Chem.Chem.Phys. 19, 32166–32178 (2017).
Battistuzzi, G., Borsari, M., Menabue, L. & Saladini, M. Amide Group Coordination to the Pb2+ Ion. Inorg. Chem. 35, 4239–4247 (1996).
Parapouli, M., Vasileiadis, A., Afendra, A. S. & Hatziloukas, E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 6, 1–31 (2020).
LESAFFRE. The yeast economy. Explore Yeast. https://www.exploreyeast.com/article/the-yeast-economy (2021).
Statista. Value of the yeast product market worldwide from 2017 to 2026. https://www.statista.com/statistics/728147/global-yeast-product-market-size/ (2021).
Jaeger, A., Arendt, E. K., Zannini, E. & Sahin, A. W. Brewer’s Spent Yeast (BSY), an Underutilized Brewing By-Product. Fermentation 6, 123 (2020).
Yang, J. et al. Nanomaterials for the Removal of Heavy Metals from Wastewater. Nanomaterials 9, 424 (2019).
Wang, C. et al. Uranium In Situ Electrolytic Deposition with a Reusable Functional Graphene‐Foam Electrode. Adv. Mater. 2102633 (2021) https://doi.org/10.1002/adma.202102633.
Wang, J. & Guo, X. Adsorption kinetic models: Physical meanings, applications, and solving methods. J. Hazard. Mater. 390, 122–156 (2020).
Wang, J. & Guo, X. Adsorption isotherm models: Classification, physical meaning, application and solving method. Chemosphere 258, 127279 (2020).
Gibbs, E., Hsu, J., Barth, K. & Goss, J. W. Characterization of the nanomechanical properties of the fission yeast (Schizosaccharomyces pombe) cell surface by atomic force microscopy. Yeast 38, 480–492 (2021).
Volle, C. B., Ferguson, M. A., Aidala, K. E., Spain, E. M. & Núñez, M. E. Quantitative Changes in the Elasticity and Adhesive Properties of Escherichia coli ZK1056 Prey Cells During Predation by Bdellovibrio bacteriovorus 109J. Langmuir 24, 8102–8110 (2008).
Stathatou, P. et al. Datasets generated from the study on trace lead removal by inactive yeast cells. figshare. Dataset. https://doi.org/10.6084/m9.figshare.19660698.v4 (2022).
Acknowledgements
We thank the MIT Koch Institute’s Robert A. Swanson (1969) Biotechnology Center for technical support, and specifically the Nanotechnology Materials Lab for assisting in TEM sample preparation and imaging. We acknowledge the MIT Materials Research Science and Engineering Center (MRSEC) for assisting with TEM imaging and ATR-FTIR analyses. We also thank the MIT Center for Environmental Health Sciences (CEHS) and in particular Dr. Bogdan Fedeles for the helpful discussions and troubleshooting concerning the ICP-MS analyses of our samples. We are grateful to Dr. Constantinos Katsimpouras from the MIT Metabolic Engineering Laboratory for assisting with the HPLC analyses. We also thank Lorena Altamirano for her assistance with biological methods and protocols. P.M.S was supported by the Bodossaki’s Foundation, Stamatis G. Mantzavinos’s Memorial Postdoctoral Scholarship. A.M. was supported by Standard Banking Group’s sponsorship to the MIT Center for Bits and Atoms (CBA). This work was also supported by MIT CBA Consortia funding.
Author information
Authors and Affiliations
Contributions
P.M.S., C.E.A., M.T., H.G., and N.G. conceived the study and designed experiments; P.M.S., C.E.A., J.G., and C.B. performed the experiments; M.T. provided technical input and helped with results interpretation; F.T., A.M, and E.M.D. contributed to data analysis and interpretation; N.P. and B.S. contributed to the final version of the manuscript and helped with results interpretation; H.G. helped in results interpretation and experimental design; All authors contributed to manuscript preparation and writing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Earth & Environment thanks Ülküye Dudu Gül, Kim Cuc Thi Nguyen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Clare Davis.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stathatou, P.M., Athanasiou, C.E., Tsezos, M. et al. Lead removal at trace concentrations from water by inactive yeast cells. Commun Earth Environ 3, 132 (2022). https://doi.org/10.1038/s43247-022-00463-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-022-00463-0
This article is cited by
-
Investigation of adsorption mechanism of Mo(VI) by baker's yeast and applicability to the uranium liquid waste treatment process
Journal of Radioanalytical and Nuclear Chemistry (2024)
-
Metallurgical pathways of lead leaching from brass
npj Materials Degradation (2023)
-
Yeast-driven valorization of agro-industrial wastewater: an overview
Environmental Monitoring and Assessment (2023)
-
Biofabricated yeast: super-soldier for detoxification of heavy metals
World Journal of Microbiology and Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.