Abstract
Immunotherapy with anti-GD2 antibodies has advanced the treatment of children with high-risk neuroblastoma, but nearly half of patients relapse, and little is known about mechanisms of resistance to anti-GD2 therapy. Here, we show that reduced GD2 expression was significantly correlated with the mesenchymal cell state in neuroblastoma and that a forced adrenergic-to-mesenchymal transition (AMT) conferred downregulation of GD2 and resistance to anti-GD2 antibody. Mechanistically, low-GD2-expressing cell lines demonstrated significantly reduced expression of the ganglioside synthesis enzyme ST8SIA1 (GD3 synthase), resulting in a bottlenecking of GD2 synthesis. Pharmacologic inhibition of EZH2 resulted in epigenetic rewiring of mesenchymal neuroblastoma cells and re-expression of ST8SIA1, restoring surface expression of GD2 and sensitivity to anti-GD2 antibody. These data identify developmental lineage as a key determinant of sensitivity to anti-GD2 based immunotherapies and credential EZH2 inhibitors for clinical testing in combination with anti-GD2 antibody to enhance outcomes for children with neuroblastoma.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Complete RNA-, ChIP- and ATAC-sequencing data are available online through the Gene Expression Omnibus (GEO) portal under project accession number GSE180516. The human neuroblastoma data were derived from the TARGET and Treehouse datasets downloaded from the Tumor Compendium v11 Public PolyA (April 2020) on UCSC Treehouse genomics website (available at https://treehousegenomics.soe.ucsc.edu/public-data). Additional human neuroblastoma data were derived from tumor sequencing data available from R2 (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Source data files for Figs. 1, 2, 3, 4, 5, 6, 7 and 8 and Extended Data Figs. 1, 2, 3, 4, 6, 7, 9 and 10 have been provided as Source Data Files. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Smith, M. A., Altekruse, S. F., Adamson, P. C., Reaman, G. H. & Seibel, N. L. Declining childhood and adolescent cancer mortality. Cancer 120, 2497–2506 (2014).
Park, J. R. et al. Effect of tandem autologous stem cell transplant vs single transplant on event-free survival in patients with high-risk neuroblastoma: a randomized clinical trial. JAMA 322, 746–755 (2019).
Yu, A. L. et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N. Engl. J. Med. 363, 1324–1334 (2010).
Yu, A. L. et al. Long-term follow-up of a phase III study of ch14.18 (dinutuximab) + cytokine immunotherapy in children with high-risk neuroblastoma: COG study ANBL0032.Clin. Cancer Res. 27, 2179–2189 (2021).
Cheung, N. K. et al. Murine anti-GD2 monoclonal antibody 3F8 combined with granulocyte-macrophage colony-stimulating factor and 13-cis-retinoic acid in high-risk patients with stage 4 neuroblastoma in first remission. J. Clin. Oncol. 30, 3264–3270 (2012).
Soldatov, R. et al. Spatiotemporal structure of cell fate decisions in murine neural crest.Science 364, eaas9536 (2019).
Boeva, V. et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 49, 1408–1413 (2017).
van Groningen, T. et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 49, 1261–1266 (2017).
Gartlgruber, M. et al. Super enhancers define regulatory subtypes and cell identity in neuroblastoma. Nat. Cancer 2, 114–128 (2020).
Berois, N. & Osinaga, E. Glycobiology of neuroblastoma: impact on tumor behavior, prognosis, and therapeutic strategies. Front. Oncol. 4, 114 (2014).
Theruvath, J. et al. Anti-GD2 synergizes with CD47 blockade to mediate tumor eradication. Nat. Med. 28, 333–344 (2022).
Yoshida, H. et al. B4GALNT1 induces angiogenesis, anchorage independence growth and motility, and promotes tumorigenesis in melanoma by induction of ganglioside GM2/GD2. Sci. Rep. 10, 1199 (2020).
Wu, Z. L., Schwartz, E., Seeger, R. & Ladisch, S. Expression of GD2 ganglioside by untreated primary human neuroblastomas. Cancer Res. 46, 440–443 (1986).
Schumacher-Kuckelkorn, R. et al. Lack of immunocytological GD2 expression on neuroblastoma cells in bone marrow at diagnosis, during treatment, and at recurrence. Pediatr. Blood Cancer 64, 46–56 (2017).
Terzic, T. et al. Expression of disialoganglioside (GD2) in neuroblastic tumors: a prognostic value for patients treated with anti-GD2 immunotherapy. Pediatr. Dev. Pathol. 21, 355–362 (2018).
Dondero, A. et al. Multiparametric flow cytometry highlights B7-H3 as a novel diagnostic/therapeutic target in GD2neg/low neuroblastoma variants.J. Immunother. Cancer 9, e002293 (2021).
Davis, T. A., Czerwinski, D. K. & Levy, R. Therapy of B-cell lymphoma with anti-CD20 antibodies can result in the loss of CD20 antigen expression. Clin. Cancer Res. 5, 611–615 (1999).
Fry, T. J. et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat. Med. 24, 20–28 (2018).
Majzner, R. G. & Mackall, C. L. Tumor antigen escape from CAR T-cell therapy. Cancer Discov. 8, 1219–1226 (2018).
Branco, F. P. et al. Loss of HER2 and disease prognosis after neoadjuvant treatment of HER2+ breast cancer. Am. J. Transl. Res. 11, 6110–6116 (2019).
Maude, S. L. et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018).
Mejstrikova, E. et al. CD19-negative relapse of pediatric B-cell precursor acute lymphoblastic leukemia following blinatumomab treatment. Blood Cancer J. 7, 659 (2017).
Keyel, M. E. & Reynolds, C. P. Spotlight on dinutuximab in the treatment of high-risk neuroblastoma: development and place in therapy. Biologics 13, 1–12 (2019).
Hirabayashi, K. et al. Dual-targeting CAR-T cells with optimal co-stimulation and metabolic fitness enhance antitumor activity and prevent escape in solid tumors. Nat. Cancer 2, 904–918 (2021).
Ross, R. A., Spengler, B. A. & Biedler, J. L. Coordinate morphological and biochemical interconversion of human neuroblastoma cells. J. Natl Cancer Inst. 71, 741–747 (1983).
Takenobu, H. et al. CD133 suppresses neuroblastoma cell differentiation via signal pathway modification. Oncogene 30, 97–105 (2011).
van Groningen, T. et al. A NOTCH feed-forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma. Nat. Commun. 10, 1530 (2019).
Ruckhaberle, E. et al. Gene expression of ceramide kinase, galactosyl ceramide synthase and ganglioside GD3 synthase is associated with prognosis in breast cancer. J. Cancer Res. Clin. Oncol. 135, 1005–1013 (2009).
Warren, A. et al. Global computational alignment of tumor and cell line transcriptional profiles. Nat. Commun. 12, 22 (2021).
Upton, K. et al. Epigenomic profiling of neuroblastoma cell lines. Sci. Data 7, 116 (2020).
Margueron, R. & Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011).
Hu, N., Strobl-Mazzulla, P. H. & Bronner, M. E. Epigenetic regulation in neural crest development. Dev. Biol. 396, 159–168 (2014).
Kailayangiri, S. et al. EZH2 inhibition in Ewing sarcoma upregulates GD2 expression for targeting with gene-modified T cells. Mol. Ther. 27, 933–946 (2019).
Wang, C. et al. EZH2 mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR. Cancer Res. 72, 315–324 (2012).
Qadeer, Z. A. et al. ATRX in-frame fusion neuroblastoma is sensitive to EZH2 inhibition via modulation of neuronal gene signatures. Cancer Cell 36, 512–527 e519 (2019).
Chen, L. et al. CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2. J. Clin. Invest. 128, 446–462 (2018).
Reppel, L. et al. Targeting disialoganglioside GD2 with chimeric antigen receptor-redirected T cells in lung cancer.J. Immunother. Cancer 10, e003897 (2022).
Shusterman, S. et al. Antitumor activity of hu14.18-IL2 in patients with relapsed/refractory neuroblastoma: a Children’s Oncology Group (COG) phase II study. J. Clin. Oncol. 28, 4969–4975 (2010).
Yin, J. et al. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc. Natl. Acad. Sci. U S A 112, 15988–15993 (2015).
Yin, Y. et al. EZH2 suppression in glioblastoma shifts microglia toward M1 phenotype in tumor microenvironment. J. Neuroinflammation 14, 220 (2017).
Zhang, X. et al. Macrophage/microglial Ezh2 facilitates autoimmune inflammation through inhibition of Socs3. J. Exp. Med. 215, 1365–1382 (2018).
Schramm, A. et al. Mutational dynamics between primary and relapse neuroblastomas. Nat Genet 47, 872–877 (2015).
Iniguez, A. B. et al. Resistance to epigenetic-targeted therapy engenders tumor cell vulnerabilities associated with enhancer remodeling. Cancer Cell 34, 922–938 (2018).
Ruan, S. & Lloyd, K. O. Glycosylation pathways in the biosynthesis of gangliosides in melanoma and neuroblastoma cells: relative glycosyltransferase levels determine ganglioside patterns. Cancer Res. 52, 5725–5731 (1992).
Lo Piccolo, M. S., Cheung, N. K. & Cheung, I. Y. GD2 synthase: a new molecular marker for detecting neuroblastoma. Cancer 92, 924–931 (2001).
Cheung, I. Y., Lo Piccolo, M. S., Collins, N., Kushner, B. H. & Cheung, N. K. Quantitation of GD2 synthase mRNA by real-time reverse transcription-polymerase chain reaction: utility in bone marrow purging of neuroblastoma by anti-GD2 antibody 3F8. Cancer 94, 3042–3048 (2002).
Jansky, S. et al. Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nat. Genet. 53, 683–693 (2021).
Dong, R. et al. Single-cell characterization of malignant phenotypes and developmental trajectories of adrenal neuroblastoma. Cancer Cell 38, 716–733 (2020).
Sequist, L. V. et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 3, 75ra26 (2011).
Aggarwal, R., Zhang, T., Small, E. J. & Armstrong, A. J. Neuroendocrine prostate cancer: subtypes, biology, and clinical outcomes. J. Natl Compr. Canc. Netw. 12, 719–726 (2014).
Neo, W. H. et al. MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J. Biol. Chem. 289, 20788–20801 (2014).
Mount, C. W. et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M+ diffuse midline gliomas. Nat. Med. 24, 572–579 (2018).
Majzner, R. G. et al. GD2-CAR T cell therapy for H3K27M-mutated diffuse midline gliomas. Nature, Epub ahead of print. PMID 35130560. PMCID: PMC8967714 (2022).
van den Bijgaart, R. J. E. et al. Combined sialic acid and histone deacetylase (HDAC) inhibitor treatment up-regulates the neuroblastoma antigen GD2. J. Biol. Chem. 294, 4437–4449 (2019).
Kroesen, M. et al. Anti-GD2 mAb and vorinostat synergize in the treatment of neuroblastoma. Oncoimmunology 5, e1164919 (2016).
Italiano, A. et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 19, 649–659 (2018).
Gounder, M. et al. Tazemetostat in advanced epithelioid sarcoma with loss of INI1/SMARCB1: an international, open-label, phase 2 basket study. Lancet Oncol. 21, 1423–1432 (2020).
Morschhauser, F. et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 21, 1433–1442 (2020).
Majzner, R. G. et al. Tuning the antigen density requirement for CAR T-cell activity. Cancer Discov. 10, 702–723 (2020).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008).
Pohl, A. & Beato, M. bwtool: a tool for bigWig files. Bioinformatics 30, 1618–1619 (2014).
Carroll, T. S., Liang, Z., Salama, R., Stark, R. & de Santiago, I. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front. Genet. 5, 75 (2014).
Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Barkal, A. A. et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 572, 392–396 (2019).
Zhang, Y. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798 (2013).
Marro, S. G. et al. Neuroligin-4 regulates excitatory synaptic transmission in human neurons. Neuron 103, 617–626 (2019).
Acknowledgements
This research was supported by the National Institutes of Health (NIH; P01 CA217959 to K.S. and R.G.M., R35 CA210030 to K.S., R01-CA227942 to C.R.B. and F30 CA232541 to B.A.H.S), the National Institute of Neurological Disorders and Stroke (R01 NS088355 to K.S., UM1 HG012076 to A.T.S. and R01-NS092597 to M.M), an NIH Director’s Pioneer Award (DP1NS111132 to M.M.), the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (M.M.), Cancer Research UK (M.M), the Waxman Family Research Fund (M.M. and A.C.G.), the Howard Hughes Medical Institute (M.M.), a Stand Up 2 Cancer (SU2C) Phillip A. Sharp Innovation in Collaboration Award (K.S., M.S. and C.S.M.) and a SU2C Convergence 2.0 grant (M.S. and C.S.M.), Alex’s Lemonade Stand (R.G.M.) and the V Foundation (R.G.M.). N.W.M. was supported by the National Cancer Institute under a Ruth L. Kirschstein National Research Service Award (F32 CA261035) and the DFCI Ungerer Fellowship award. R.G.M. is the Taube Distinguished Scholar for Pediatric Immunotherapy at Stanford University School of Medicine. A.T.S. was supported by a Technology Impact Award from the Cancer Research Institute, a Career Award for Medical Scientists from the Burroughs Wellcome Fund and a Pew-Stewart Scholars Award. M.M.M. was supported by the Deutsche Forschungsgemeinshaft (MA 8492/1-1). D.A.S was supported by the Internationales Forschungsstipendium from the Medical Curriculum Munich and the Biomedical Education Program. We thank members of the Program Project Grant and New Approaches to Neuroblastoma Therapy group for their thoughtful comments and suggestions. We thank M. Kararoudi and D. Lee at Nationwide Children’s Hospital for their advice and helpfulness with establishing NK cell culture methods.
Author information
Authors and Affiliations
Contributions
N.W.M., M.H., K.S. and R.G.M. conceived of the project. N.W.M., M.H., N.D., D.A.S., A.C.G., M.M.M., M.L., N.L., W.Y., M.C.R., B.A.H.S., C.M., R.M., A.S., K.S. and R.G.M. generated data. M.S. and C.M. provided NK cell cultures and guidance. N.W.M., M.H., N.D., H.A., A.C.G., M.M.M., J.A.B., B.A.H.S., K.S. and R.G.M. analyzed data. N.W.M., M.H., N.D., A.S.C., M.M.M., M.L., B.A.H.S., M.C.R., M.W., C.R.B., M.M., C.M., R.M., A.T.S., K.S. and R.G.M. helped design experiments. N.D., A.L.R., A.S.C. an D.K. planned and executed in vivo studies. N.W.M., G.A., J.A.B., K.N.R. and A.T.S. provided bioinformatics analysis. N.W.M., K.S. and R.G.M. wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
K.S. consults for and has stock options in Auron Therapeutics, received grant funding from Novartis, served as an advisor for KronosBio and consulted for AstraZeneca and Bristol Meyers Squibb on topics unrelated to this manuscript. R.G.M. is a co-founder of and holds equity in Syncopation Life Sciences and Link Cell Therapies and is a consultant for Lyell Immunopharma, Innvervate Radiopharmaceuticals, NKarta, Immunai, Arovella Therapeutics, Zai lab and Aptorum Group. A.T.S. is a co-founder of Immunai and Cartography Biosciences and receives research funding from Arsenal Biosciences, Merck Research Laboratories and Allogene Therapeutics. C.R.B. is a co-founder of Redwood Biosciences (a subsidiary of Catalent), Enable Biosciences, Palleon Pharmaceuticals, InterVenn Bio, Lycia Therapeutics, OliLux Biosciences, Grace Science and Virsti Therapeutics. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks Christian Capitini and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Low GD2 expression is correlated with developmental lineage in neuroblastoma cell culture models.
a, Bar plot showing the percentage of GD2 + cells for each cell line from Fig. 1a (n = 12 GD2-high, n = 11 GD2-low). Data are shown as mean ± s.d. b, Scatterplot comparing adrenergic (ADRN) and mesenchymal (MES) composite scores for neuroblastoma cell lines in the CCLE. ADRN and MES scores were calculated based on average log2(TPM + 1) expression of all genes within each gene set. Mesenchymal cell lines were called based on MES score ≥ 4.1 and ADRN score ≤ 5. c, Heatmap showing median-center, Zz-score normalized RNA sequencing for adrenergic (ADRN, green) and mesenchymal (MES, purple) genes in all neuroblastoma cell lines with RNA-sequencing data available in the CCLE. d, Semi-supervised, hierarchical heatmap showing Zz-score normalized expression data for mesenchymal (purple) and adrenergic (green) gene sets in cell lines analyzed by differential gene expression analysis and for which GD2 status is shown in Fig. 1a. e, Western blot showing expression of mesenchymal markers Vimentin, fibronectin, TAZ and YAP1 and adrenergic marker PHOX2B in SH-EP and SH-SY5Y cell lines. GAPDH is shown as a control. Note that the GAPDH panel is the same bands are shown in Fig. 4a. f, Flow cytometry panels demonstrating CD16 staining in two healthy, donor-derived NK cell cultures. g, Cell viability for SH-SY5Y or SH-EP cell lines co-cultured with NK cells at an E:T ratio of 1:2 for 48 h and in the presence or absence of 1 μg/mL dinutuximab (n = 3 samples per treatment group). Data are shown mean ± s.d. Significance determined by one-way ANOVA and Tukey’s post-hoc test. ns = not significant. Representative data from western blots were confirmed in two independent experiments.
Extended Data Fig. 2 GD2 density influences response to anti-GD2 antibody.
a, Parental Kelly cells were sorted based on GD2 expression into Kelly-GD2low (red) or Kelly-GD2high (black) isogenic cell lines. b, Kelly-GD2low or Kelly-GD2high were co-cultured with blood-derived macrophages from three healthy donors and measured for phagocytosis in presence or absence of anti-GD2. Data shown is phagocytosis with dinutuximab and normalized to the control condition for that cell line. Triplicates for all three donor cultures were combined. Data are shown mean ± s.d. Significance was determined by two-tailed Mann-Whitney U test. c, Cell viability for Kelly-GD2low or Kelly-GD2high cell lines co-cultured with NK cells at an E:T ratio of 1:2 for 48 h in the presence or absence of 1 μg/mL dinutuximab (n = 3 samples per treatment group). Data are shown mean ± s.d. Significance determined by one-way ANOVA and Tukey’s post-hoc test. d, Flow cytometry for Kelly-GD2low and Kelly-GD2high cells for mesenchymal cell marker CD133 (PROM1). ns = not significant. Representative data from flow cytometry were confirmed in two independent experiments.
Extended Data Fig. 3 Induced Adrenergic-to-Mesenchymal Transition is associated with GD2 downregulation.
a, Gene Set Enrichment Analysis was performed with the neuroblastoma-specific adrenergic or mesenchymal gene signatures for RNA sequencing from vehicle or doxycycline-treated SK-N-BE(2)-tetON-PRRX1 and KP-N-YN-tetON-PRRX1 cell lines. q-values are shown. b, Cell viability for KP-N-YN induced AMT models co-cultured with NK cells at an E:T ratio of 1:2 in the presence or absence of 1 μg/mL dinutuximab for 8 h (n = 4 samples per treatment group). Data are shown as mean ± s.d. Significance determined by one-way ANOVA and Tukey’s post-hoc test. c, Western blot showing mesenchymal (Vimentin, TAZ, NOTCH3IC) or adrenergic (PHOX2A) markers in parental or NOTCH3IC-expressing SK-N-BE(2) cells. GAPDH is included as a control. d, Representative micrographs (20X) of parental, PRRX1-, or NOTCH3IC-expressing SK-N-BE(2) cells. e, Flow cytometry analysis of GD2 for NOTCH3IC-overexpressing SK-N-BE(2) cells. ns = not significant. Representative data from flow cytometry and western blots were confirmed in two independent experiments.
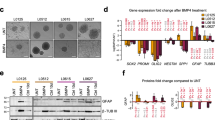
Extended Data Fig. 4 Low ST8SIA1 expression correlates with low surface GD2 and mesenchymal features.
a, Schematic showing the complete ganglioside synthesis pathway. Enzymes responsible for conversion of each ganglioside are labeled in bold and branch points are colored. b, Parental SH-EP (top) or CHLA-255 (bottom) cell lines were sorted based on GD2 expression into GD2low (red) or GD2high (black) isogenic cell lines, respectively. c, qPCR analysis comparing expression for ST8SIA1, B4GALNT1 and ST3GAL5 in the SH-EP-GD2high and SH-EP-GD2low (top) or CHLA-255-GD2high and CHLA-255-GD2low (bottom) isogenic cell line pairs. Data derived from a single experiment with 4 technical replicates, experiment was completed once. d, Cell viability for Kelly-GD2low and NB-SD with or without GD3 synthase overexpression and co-cultured with NK cells at an E:T ratio of 1:2 and in the presence of absence of 1 μg/mL dinutuximab for 8 h (n = 4 samples per treatment group). Data are shown as mean ± s.d. Significance determined by one-way ANOVA and Tukey’s post-hoc test. e, Flow cytometry plot showing GD2 expression in Kelly-GD2low, SK-N-AS and NB-SD cell lines with or without constitutive expression of ST3GAL5. f, qPCR analysis measuring ST8SIA1 expression in parental or SK-N-BE(2)-NOTCH3IC cells. Data derived from a single experiment with 4 technical replicates, experiment was repeated twice. ns = not significant. Representative data from flow cytometry were confirmed in two independent experiments.
Extended Data Fig. 5 Neuroblastoma tumors with mesenchymal properties express reduced GD3 synthase.
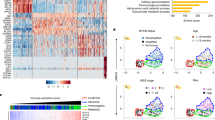
UMAP 2D projection plot showing integrated global gene expression for tumors (Treehouse/TARGET) and cell lines (CCLE) from 43 tumor lineages from Celligner. The neuroblastoma lineage is highlighted within the red box.
Extended Data Fig. 6 EZH2 inhibition restores GD2 expression by reversal of epigenetic silencing of GD3 synthase.
a, Flow Cytometry analysis measuring GD2 expression in LAN-5 cells. b, ChIP-seq tracks showing active histone mark H3K27ac and the repressive histone mark H3K27me3 at the ST8SIA1 locus in LAN-5 (GD2-high), NB-69 (GD2-high), SK-N-AS (GD2-low) and SK-N-BE(2)C (GD2-low) cells from GSE138314. c, Flow cytometry panel for GD2 expression in SK-N-AS cells treated with increasing concentrations of tazemetostat. d, Flow cytometry plot showing GD2 expression for CHLA-255-GD2low cells treated for 21 days with 1 μM tazemetostat. e, Mean fluorescence intensity for GD2 in the flow cytometry data shown in panel d (n = 3 samples). Data are shown mean ± s.d. Significance was determined by two-tailed Student’s t-test. f, Flow cytometry panels showing GD2 expression in Ewing sarcoma cell lines SK-PN-DW, EW-8 and TC-32 treated for 21 days with 1 μM tazemetostat or the small cell lung cancer cell lines H-69 and H-82 cell lines treated for 14 days with 1 μM tazemetostat. g, Flow cytometry for GD2 expression in SK-N-AS cells treated for 21 days with 1 μM tazemetostat or treated for 21 days and given a 1-month drug holiday. Representative data from flow cytometry and western blots were confirmed in two independent experiments.
Extended Data Fig. 7 Integrated analysis of RNA-, ChIP- and ATAC-sequencing following EZH2 inhibition in SK-N-AS cells.
a, Heatmap showing H3K27me3 ChIP-seq signal at all detected H3K27me3 peaks (88,858). Heatmap is separated into genomic regions with H3K27me3 that are decreased (82,122), unchanged (2,862) or increased (3,874) with 21-day treatment with 1 μM tazemetostat in SK-N-AS cells. PC = peak center, ns = not changed, incr = increased. b, Signal enrichment profile plot showing average H3K27me3 enrichment signal for vehicle or tazemetostat treatment groups. Average signal is calculated from 88,858 H3K27me3 peaks. Average signal was normalized to background signal. Values are shown ±5 kb from the peak center. Significance was determined by two-tailed Student’s t-test for the area-under-the-curve. c, Scatterplot correlating the log2 fold change in H3K27me3 area-under-the-curve (AUC) signal with the log2 fold change in RNA expression. The number of significant genes (fold change of RNA ≥ 2 and H3K27me3 ∆ log2 AUC ≤ -0.5) in each quadrant are indicated. Significance for the number of genes within a quadrant was determined by two-tailed Fisher’s exact test. d, Scatterplot correlating the log2 fold change in ATAC area-under-the-curve (AUC) with the log2 fold change in RNA expression. The number of significant genes (fold change of RNA ≥ 2 and H3K27me3 ∆ log2 AUC ≥ 0.5) in each quadrant are indicated. Significance for the number of genes within a quadrant was determined by two-tailed Fisher’s exact test. e, Venn diagram showing the number of overlapping genes for which H3K27me3 signal was lost, ATAC-seq signal was gained, and RNA expression increased. ***P < 0.001.
Extended Data Fig. 8 Pathways enriched in epigenetically regulated genes from integrated RNA-, ChIP- and ATAC-sequencing following EZH2 inhibition.
Dot plot showing the significant overlap of the 575 genes of interest shown in Extended Data Fig. 7e with C2 and C5 MSigDB libraries. Categories were clustered into the top six most similar pathways. Nervous system development/differentiation-related pathways are indicated in red. Dot size indicates the extent of gene overlap with the indicated gene sets. All gene sets have an FDR of ≤ 0.05 and were calculated by one-tailed Fisher’s exact test based on the hypergeometric distribution of the overlapping 575 genes.
Extended Data Fig. 9 EZH2 inhibition increases GD2 density and response to anti-GD2 in multiple models in vivo.
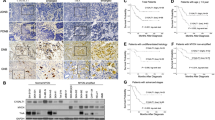
a, Kelly parental cells were injected into the tail vein of NSG mice (n = 4 vehicle, n = 5 tazemetostat) and treated with 350 mg/kg tazemetostat twice daily or control. Flow cytometry panel showing GD2 expression in representative tumors measured at tumor endpoint. b, Quantification of mean fluorescence intensity for GD2 expression for all parental Kelly tumors treated with control (n = 4) or tazemetostat (n = 5). Data are shown as mean ± s.d. Significance was determined by two-tailed Student’s t-test. c, Representative flow cytometry panel for GD2 from MG63.3 osteosarcoma cells orthotopically injected into the hind leg of NSG mice (n = 4 vehicle, n = 5 tazemetostat) and treated with 350 mg/kg tazemetostat twice daily. d, Quantification of mean fluorescence intensity for GD2 expression in all MG63.3 tumors treated with or without tazemetostat. Data are shown as mean ± s.d. Significance was determined by two-tailed Student’s t-test. e, Flow cytometry panel for SK-N-AS cells injected into the tail vein of NSG mice (n = 5 per treatment group) and treated with 300 μg dinutuximab three times a week or 500 mg/kg tazemetostat twice daily alone or in combination. Note that one representative tumor from untreated and tazemetostat only groups were shown in Fig. 7A. Representative data from flow cytometry are shown for the biological replicates presented in panels a and c.
Extended Data Fig. 10 EZH2 inhibition increases anti-GD2 response in vivo.
a, Bar plots showing the population of macrophage (MP), granulocyte (Gran), monocyte (Mono) and dendritic cell (DC) populations in each treatment arm (n = 5 per arm) as determined by flow cytometry. Population percentages were determined by the following markers within CD45 + cells: macrophages: CD11b + / F4/80 + ; granulocytes: CD11b + / Ly6G + ; monocytes: CD11b + / Ly6C + ; dendritic cells: CD11c + / MHC-II + . Data are shown as mean ± s.d. Significance was determined by one-way ANOVA and Tukey’s post-hoc test. b, Bar plots showing the percent of M1 macrophages (CD86 + / MHC-II (I-A/I-E) + ) or M2 macrophages (CD163 + /CD206 + ) as a total of the macrophage population (CD11b + / F480 + ) (n = 5 per arm). Data are shown as mean ± s.d. Significance was determined by two-tailed Student’s t-test. c, Flow cytometry showing representative GD2 expression in treatment naïve or tazemetostat pretreated tumors treated with either dinutuximab or anti-GD2 CAR T cells in vivo. d,e, Immunofluorescence staining for GD2 or neuronal marker MAP2 on vehicle or tazemetostat-treated, passaged primary human cortical cells (d) or inducible embryonic stem cells (e). f, Human embryonic stem cells were differentiated into induced neurons and treated for 14 days with 1 μM tazemetostat. GD2 was measured by flow cytometry. ns = not significant. Representative data from flow cytometry were confirmed in two independent experiments.
Supplementary information
Supplementary Information
Supplemental Figures 1–7 showing representative gating strategies for flow cytometry panels.
Supplementary Table 1
GSEA for significant Hallmark gene sets between GD2-high and GD2-low cell lines. Supplementary Table 2. Genes correlating with percentage of GD2+ cells in neuroblastoma cell lines. Supplementary Table 3. Summary master file for integrated RNA, ChIP and ATAC sequencing in vehicle or tazemetostat-treated SK-N-AS cells. P values were calculated by DESeq2 on apeglm settings and multiple corrections testing using false discovery rate. Supplementary Table 4. Extended list of gene ontology categories enriched in the 575 overlapping genes shown in Extended Data Figs. 7e and 8. Significance and adjusted P values were determined by hypergeometric distribution analysis using one-tailed Fisher’s exact test for values greater than the contingency table. Supplementary Table 5. Reagents used for studies.
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 2
Uncropped western blots
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 4
Uncropped western blots
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 6
Statistical Source Data
Source Data Fig. 6
Uncropped western blots
Source Data Fig. 7
Statistical Source Data
Source Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 1
Uncropped western blots
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 3
Uncropped western blots
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 7
Statistical Source Data
Source Data Extended Data Fig. 9
Statistical Source Data
Source Data Extended Data Fig. 10
Statistical Source Data
Rights and permissions
About this article
Cite this article
Mabe, N.W., Huang, M., Dalton, G.N. et al. Transition to a mesenchymal state in neuroblastoma confers resistance to anti-GD2 antibody via reduced expression of ST8SIA1. Nat Cancer 3, 976–993 (2022). https://doi.org/10.1038/s43018-022-00405-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-022-00405-x
This article is cited by
-
Noninvasive serum N-glycans associated with ovarian cancer diagnosis and precancerous lesion prediction
Journal of Ovarian Research (2024)
-
A sialyltransferases-related gene signature serves as a potential predictor of prognosis and therapeutic response for bladder cancer
European Journal of Medical Research (2023)
-
Identification of the molecular subtypes and construction of risk models in neuroblastoma
Scientific Reports (2023)