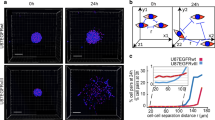
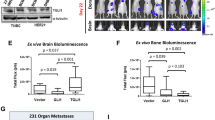
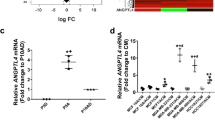
Abstract
Although dormancy is thought to play a key role in the metastasis of breast tumor cells to the brain, our knowledge of the molecular mechanisms regulating disseminated tumor cell (DTC) dormancy in this organ is limited. Here using serial intravital imaging of dormant and metastatic triple-negative breast cancer lines, we identify escape from the single-cell or micrometastatic state as the rate-limiting step towards brain metastasis. We show that every DTC occupies a vascular niche, with quiescent DTCs residing on astrocyte endfeet. At these sites, astrocyte-deposited laminin-211 drives DTC quiescence by inducing the dystroglycan receptor to associate with yes-associated protein, thereby sequestering it from the nucleus and preventing its prometastatic functions. These findings identify a brain-specific mechanism of DTC dormancy and highlight the need for a more thorough understanding of tumor dormancy to develop therapeutic approaches that prevent brain metastasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Single-cell RNA sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession code GSE152818. Source data are provided with this paper. Source data for Figs. 1–5 and 7 and Extended Data Figs. 2–8 have been provided as Source data files. All other data supporting the findings of this study are available from the corresponding authors on request.
References
Steeg, P. S., Camphausen, K. A. & Smith, Q. R. Brain metastases as preventive and therapeutic targets. Nat. Rev. Cancer 11, 352–363 (2011).
Quail, D. F. & Joyce, J. A. The microenvironmental landscape of brain tumors. Cancer Cell 31, 326–341 (2017).
Zimmer, A. S. et al. Temozolomide in secondary prevention of HER2-positive breast cancer brain metastases. Future Oncol. 16, 899–909 (2020).
Lorger, M. & Felding-Habermann, B. Capturing changes in the brain microenvironment during initial steps of breast cancer brain metastasis. Am. J. Pathol. 176, 2958–2971 (2010).
Bos, P. D. et al. Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009 (2009).
Joyce, J. A. & Pollard, J. W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 9, 239–252 (2009).
Witzel, I., Oliveira-Ferrer, L., Pantel, K., Muller, V. & Wikman, H. Breast cancer brain metastases: biology and new clinical perspectives. Breast Cancer Res. 18, 8 (2016).
Paget, S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 8, 98–101 (1989).
Noltenius, C. & Noltenius, H. Dormant tumor cells in liver and brain. An autopsy study on metastasizing tumors. Pathol. Res. Pract. 179, 504–511 (1985).
Heyn, C. et al. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn. Reson. Med. 56, 1001–1010 (2006).
Ghajar, C. M. et al. The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 15, 807–817 (2013).
Carlson, P. et al. Targeting the perivascular niche sensitizes disseminated tumour cells to chemotherapy. Nat. Cell Biol. 21, 238–250 (2019).
Price, T. T. et al. Dormant breast cancer micrometastases reside in specific bone marrow niches that regulate their transit to and from bone. Sci. Transl. Med. 8, 340ra373 (2016).
Kienast, Y. et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 16, 116–122 (2010).
Stoletov, K. et al. Role of connexins in metastatic breast cancer and melanoma brain colonization. J. Cell Sci. 126, 904–913 (2013).
Carbonell, W. S., Ansorge, O., Sibson, N. & Muschel, R. The vascular basement membrane as ‘soil’ in brain metastasis. PLoS ONE 4, e5857 (2009).
Er, E. E. et al. Pericyte-like spreading by disseminated cancer cells activates YAP and MRTF for metastatic colonization. Nat. Cell Biol. 20, 966–978 (2018).
Janzer, R. C. & Raff, M. C. Astrocytes induce blood-brain barrier properties in endothelial cells. Nature 325, 253–257 (1987).
Armulik, A. et al. Pericytes regulate the blood-brain barrier. Nature 468, 557–561 (2010).
Liddelow, S. & Barres, B. SnapShot: astrocytes in health and disease. Cell 162, 1170 (2015).
Ridet, J. L., Malhotra, S. K., Privat, A. & Gage, F. H. Reactive astrocytes: cellular and molecular cues to biological function. Trends Neurosci. 20, 570–577 (1997).
Contreras-Zarate, M. J. et al. Estradiol induces BDNF/TrkB signaling in triple-negative breast cancer to promote brain metastases. Oncogene 38, 4685–4699 (2019).
Marchetti, D., Li, J. & Shen, R. Astrocytes contribute to the brain-metastatic specificity of melanoma cells by producing heparanase. Cancer Res. 60, 4767–4770 (2000).
Doron, H. et al. Inflammatory activation of astrocytes facilitates melanoma brain tropism via the CXCL10-CXCR3 signaling axis. Cell Rep. 28, 1785–1798 (2019).
Zhang, L. et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527, 100–104 (2015).
Palmieri, D. et al. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain. Cancer Res. 67, 4190–4198 (2007).
Abbott, N. J., Ronnback, L. & Hansson, E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 7, 41–53 (2006).
Nagelhus, E. A. & Ottersen, O. P. Physiological roles of aquaporin-4 in brain. Physiol. Rev. 93, 1543–1562 (2013).
Sixt, M. et al. Endothelial cell laminin isoforms, laminins 8 and 10, play decisive roles in T cell recruitment across the blood-brain barrier in experimental autoimmune encephalomyelitis. J. Cell Biol. 153, 933–946 (2001).
Agrawal, S. et al. Dystroglycan is selectively cleaved at the parenchymal basement membrane at sites of leukocyte extravasation in experimental autoimmune encephalomyelitis. J. Exp. Med. 203, 1007–1019 (2006).
Menezes, M. J. et al. The extracellular matrix protein laminin alpha2 regulates the maturation and function of the blood-brain barrier. J. Neurosci. 34, 15260–15280 (2014).
Berzin, T. M. et al. Agrin and microvascular damage in Alzheimer’s disease. Neurobiol. Aging 21, 349–355 (2000).
Willis, R. A. The Spread of Tumours in the Human Body (Butterworth & Co., 1952).
Wasilewski, D., Priego, N., Fustero-Torre, C. & Valiente, M. Reactive astrocytes in brain metastasis. Front. Oncol. 7, 298 (2017).
Seandel, M. et al. Generation of a functional and durable vascular niche by the adenoviral E4ORF1 gene. Proc. Natl Acad. Sci. USA 105, 19288–19293 (2008).
Vanlandewijck, M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018).
Liu, J. et al. A human cell type similar to murine central nervous system perivascular fibroblasts. Exp. Cell Res. 402, 112576 (2021).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Yurchenco, P. D. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 3, a004911 (2011).
Cimino, P. J. Jr. & Perrin, R. J. Mammaglobin-A immunohistochemistry in primary central nervous system neoplasms and intracranial metastatic breast carcinoma. Appl. Immunohistochem. Mol. Morphol. 22, 442–448 (2014).
Yao, Y., Chen, Z. L., Norris, E. H. & Strickland, S. Astrocytic laminin regulates pericyte differentiation and maintains blood brain barrier integrity. Nat. Commun. 5, 3413 (2014).
Chen, Z. L. et al. Ablation of astrocytic laminin impairs vascular smooth muscle cell function and leads to hemorrhagic stroke. J. Cell Biol. 202, 381–395 (2013).
Cheng, Y. S., Champliaud, M. F., Burgeson, R. E., Marinkovich, M. P. & Yurchenco, P. D. Self-assembly of laminin isoforms. J. Biol. Chem. 272, 31525–31532 (1997).
Grzelak CA. et al. Elimination of fluorescent protein immunogenicity permits modeling of metastasis in immune-competent settings. Cancer Cell (in the press).
Campbell, K. P. & Kahl, S. D. Association of dystrophin and an integral membrane glycoprotein. Nature 338, 259–262 (1989).
Weaver, V. M. et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 137, 231–245 (1997).
Gumbiner, B. M. & Kim, N. G. The Hippo-YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 127, 709–717 (2014).
Wang, Y. et al. Comprehensive molecular characterization of the Hippo signaling pathway in cancer. Cell Rep. 25, 1304–1317 (2018).
Cheung, T. H. & Rando, T. A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 14, 329–340 (2013).
Lin, Z. et al. Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity. eLife 8, e49439 (2019).
Morikawa, Y., Heallen, T., Leach, J., Xiao, Y. & Martin, J. F. Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature 547, 227–231 (2017).
Fredriksson, S. et al. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 (2002).
Zhao, B. et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007).
Zhao, B., Li, L., Tumaneng, K., Wang, C. Y. & Guan, K. L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24, 72–85 (2010).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008).
Albrengues, J. et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science 361, eaa04227 (2018).
Ghajar, C. M. Metastasis prevention by targeting the dormant niche. Nat. Rev. Cancer 15, 238–247 (2015).
Martin, P. T. Mechanisms of disease: congenital muscular dystrophies-glycosylation takes center stage. Nat. Clin. Pract. Neurol. 2, 222–230 (2006).
Tronche, F. et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 (1999).
Chen, Z. L. & Strickland, S. Laminin gamma1 is critical for Schwann cell differentiation, axon myelination, and regeneration in the peripheral nerve. J. Cell Biol. 163, 889–899 (2003).
Saederup, N. et al. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS ONE 5, e13693 (2010).
Briand, P., Nielsen, K. V., Madsen, M. W. & Petersen, O. W. Trisomy 7p and malignant transformation of human breast epithelial cells following epidermal growth factor withdrawal. Cancer Res. 56, 2039–2044 (1996).
Cailleau, R., Olive, M. & Cruciger, Q. V. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro 14, 911–915 (1978).
Yoneda, T., Williams, P. J., Hiraga, T., Niewolna, M. & Nishimura, R. A bone-seeking clone exhibits different biological properties from the MDA-MB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. J. Bone Miner. Res. 16, 1486–1495 (2001).
Harper, K. L. et al. Mechanism of early dissemination and metastasis in Her2(+) mammary cancer. Nature 540, 588–592 (2016).
She, W. et al. Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development 140, 4008–4019 (2013).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Tarashansky, A. J., Xue, Y., Li, P., Quake, S. R. & Wang, B. Self-assembling manifolds in single-cell RNA sequencing data. eLife 8, e48994 (2019).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Yang, X. et al. A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 (2011).
Oki, T. et al. A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0-G1 transition. Sci. Rep. 4, 4012 (2014).
Li, P. et al. alphaE-catenin inhibits a Src-YAP1 oncogenic module that couples tyrosine kinases and the effector of Hippo signaling pathway. Genes Dev. 30, 798–811 (2016).
Ganesh, K. et al. L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat. Cancer 1, 28–45 (2020).
Acknowledgements
We thank S. Beronja (FHCRC) for reading the manuscript and providing critical feedback, and the entire Ghajar laboratory for helpful discussions. We thank P. Steeg (NCI) for generously providing us with the MDA-MB-231-BR7 cell line; P. Paddison (FHCRC) for ECM overexpression constructs; V. Vasioukhin (FHCRC) for YAP mutant overexpression constructs; W. Stallcup (Sanford Burnham) for generously providing us with the PDGFRβ antibody; M. Ruegg (University of Basel) for providing agrin cDNA and antibody; J. Vazquez Lopez and E. Schweitzer (FHCRC) for assistance with two-photon microscopy; F. Szulzewsky (FHCRC) for providing Cx3cr1-GFP;CCR2-RFP reporter mice; and S. Strickland (Rockefeller University) for contributing Lamγ1 floxed mice through The Jackson Laboratory. This study was catalyzed by start-up funds provided by the FHCRC, and supported to its completion by funding from NIH/NCI (no. R01 CA252874, to C.M.G.), a Physical Sciences Oncology Project Grant from NIH/NCI (no. U54CA193461-01, to E.C.H.), awards from the Department of Defense (DoD) Breast Cancer Research Program (BCRP, no. W841XWH-15-1-0201 to C.M.G. and K.C.H. and no. W81XWH-19-1-0076 to C.M.G.) and by the Comparative Medicine, Experimental Histopathology and Genomics Shared Resources of the Fred Hutch/University of Washington Cancer Consortium (no. P30 CA015704). Rapid-autopsy specimen collection at Huntsman Cancer Institute was supported by Huntsman Cancer Foundation and Halt Cancer at X. J.D. was supported by a Postdoctoral Breakthrough award (no. W81XWH-18-1-0028) by the DoD BCRP. A.R.L. was supported by a fellowship from NCI (no. F99CA234840-02). C.A.G. was supported by a postdoctoral fellowship from the Susan G. Komen Foundation. L.P. was supported by Postdoc.Mobility fellowships (nos. 165389 and 177917) from the Swiss National Science Foundation. F.W. received funding from Deutsche Krebshilfe (German Cancer Aid), Priority Program ‘Translational Oncology’ no. 70112507, ‘Preventive strategies against brain metastases’. Some autopsy materials used in this study were obtained from the University of Washington Neuropathology Core, which is supported by the Alzheimer’s Disease Research Center (no. AG05136), the Adult Changes in Thought Study (no. AG006781) and Morris K. Udall Center of Excellence for Parkinson’s Disease Research (no. NS062684). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
J.D. made fundamental intellectual contributions, performed experiments and analyzed and interpreted data. P.J.C., A.L.W., S.W. and A.U. were instrumental in the acquisition and analysis of human tissue. C.A.G., A.R.L., S.W., A.U., L.T.S., L.P. and V.S. performed experiments and/or analyzed data. A.R.L. helped with scientific illustration. A.B. and K.C.H. conducted ECM-targeted mass spectrometry and analyzed and interpreted the resulting data. J.H.B. helped with design of the single-cell RNA sequencing experiment. A.L. and N.C. conducted single-cell RNA sequencing library preparation. K.H.G. and S.K. analyzed and interpreted the resulting data. M.J.B., D.L., F.W. and E.C.H. provided key scientific insight. F.W. provided instruction for intravital imaging experiments. C.M.G. conceived of the study, conducted experiments and analyzed and interpreted data. C.M.G. and J.D. wrote the manuscript. All authors read and provided feedback on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cancer thanks Christoph Klein and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Intravital imaging reveals micrometastatic progression and regression of DTCs in brain.
Serial intravital imaging revealed several potential fates for eGFP-T4-2 breast cancer DTCs which reached the brain: a) DTCs progress to micrometastatic lesions but regress and die by day 20 post-intracardiac inoculation; b) proliferate then regress by day 23, but continue to persist as a single cell through day 39; and c) steadily progress to form stable micrometastases. These behaviors were also observed in a second model using MDA-MB-231 cells: d) steady progression to form micrometastases by day 14, which continue to grow along vasculature through day 43; e) death/disappearance of micrometastasis by day 29. Representative images of n = 16 cells over 6 mice (T4-2) and n = 17 cells over 5 mice (MDA-MB-231) over the course of 4 to 7 weeks post-injection were tracked for regression (a,b,e) or progression (c,d). Scale bar = 40 µm.
Extended Data Fig. 2 GFAP staining confirms that astrocytes are stripped from vessels upon activation of micrometastases.
a) Representative Z-projections of disseminated eGFP-T4-2 cells and astrocyte (GFAP+, cyan) coverage of DTC-associated vessels in the brains of NOD SCID mice across four stages of brain metastatic progression, 8 weeks following intracardiac inoculation. Yellow arrows indicate Ki67+ tumour cells (see inset for CD31 and GFAP channels only). Scale bar = 20 µm. From left to right, images are representative of n = 12, 5, 6 and 12 independent lesions imaged across n = 3 mice. b) Quantification of astrocyte vessel coverage across metastatic progression. P-values calculated for n = 35 lesions across 3 mice by one-way ANOVA and Turkey’s post hoc test. Centre line represents the mean, and error bars the s.e.m.
Extended Data Fig. 3 Astrocytes suppress breast tumour cell outgrowth in a model of the brain’s perivascular niche.
a) Representative IF images of MDA-MB-231 cell outgrowth on F vs. F + E vs. F + E + A co-cultures 10 days post-seeding. Scale bar = 500 μm. Inset: Enlarged images to illustrate Ki67 status of MDA-MB-231 cells. Scale bar = 100 μm. Images representative of 5 wells scanned per condition across n = 4 biologically independent experiments. b) Tumour cell area fraction of YFP-MDA-MB-231 cells at day 10, normalized by tumour area fraction post-seeding, then normalized to the mean of HBAF (F) condition replicates to correct for the variations between independent replicates. *P = 0.026, **P = 0.003 for n = 6 co-culture wells analyzed per condition over 4 independent experiments, by one-way ANOVA and Tukey’s post hoc test. c) Fraction of Ki67-negative tumour cell clusters. *P = 0.012 for F + E + A vs. F; P = 0.023 for F + E vs. F + E + A, for n = 6 wells of co-cultures analyzed per condition, 4 independent experiments, by one-way ANOVA and Tukey’s post hoc test. d, e) Tumour cell area fraction of YFP-T4-2 cells at day 10, normalized by tumour area fraction post-seeding, as a function of (d) increasing endothelial cell (EC) seeding density (while holding astrocyte number constant) or (e) increasing astrocyte seeding density (while holding EC number constant). Representative data from one experiment with n = 5 technical replicates, experiments were conducted twice with similar results. f) Quantification of normalized tumour area fraction of YFP-T4-2 cells outgrowth on HBAF (F), endothelia (E), astrocyte (A), all pairwise combinations, and F + E + A co-cultures, 10-days after seeding. NS denotes no significance (P > 0.37); *P < 0.05 compared to F condition, for n = 5 sets of co-cultures analyzed per condition over 4 independent experiments, by one-way ANOVA and Tukey’s post hoc test. g) Normalized tumour area fraction 10 days after addition of conditioned media diluted 1:1 with fresh medium. NS denotes no significance (P > 0.47), for n = 6 wells per co-culture condition, analyzed for two independent experiments by one-way ANOVA and Tukey’s post hoc test. h) Representative 96-well tile scans of YFP-T4-2 cells grown on HBAF, in the presence of indicated concentrations of Laminin (LN)-211, LN-111, LN-411 or LN-511, 10 days post-seeding. Images are representative of n = 5 sets of co-cultures analyzed per condition across n = 3 biologically independent experiments. Scale bar = 1 mm. All data are presented as mean values + /− SEM.
Extended Data Fig. 4 Lama2 is predominantly expressed by astrocytes, and is stripped from vessels over the course of metastatic progression.
a) Transcript expression levels for Lama2, Agrin, Col18A1, and Vcan for astrocytes, neurons, oligodendrocyte progenitor cells (OPC), newly formed oligodendrocytes, myelinating oligodendrocytes, microglia and endothelial cells, immunopanned from adult murine brains and sequenced by RNAseq. Data are reported as FPKM, and were generated by querying brainrnaseq.org. b, c) Representative Z-projections of disseminated (b) mCherry-MDA-MB-231 cells (false colored green for consistency) or (c) eGFP-T4-2 cells in brains of NOD-SCID mice ~8 weeks after intracardiac inoculation, demonstrating that stripping of (b) Laminin-α2 but not of (c) other laminins coincides with metastatic progression. Arrows indicate Ki67+ nuclei in a micrometastatic cluster. Scale bar = 20 μm for three leftmost panels and 100 μm for MACRO-Metastasis. Images representative of n = 6 sections per mouse acquired for n = 3 mice per condition. d) Representative IF staining for Laminin-α2 (left panels) and pan-laminin (right panels) to assess vessel coverage in brain-tropic MDA-MB-231-BR7-derived metastatic lesions. Scale bar = 20 µm. From left to right, images representative of n = 13, 10, 12 and 16 independent fields obtained across n = 4 mice. e) Mean fluorescent intensity of Laminin-α2 and pan-laminin within macrometastatic lesions (‘Tumour’) and lesion-adjacent regions (‘Adjacent’). ***P < 0.0001 when comparing tumour to adjacent tissue, for n = 51 total lesions across 4 mice by unpaired, two-tailed t-test. Data are presented as mean values + /− SEM.
Extended Data Fig. 5 Further characterization of Nestin-drive KO of lamc1.
a) Representative IF staining of AQP4 and CD31 from Nestin-Cre, Lamγ1flox/flox conditional knockout (N-γ1-KO) mice and littermate controls. Samples from 3 different mice shown. Scale bar = 30 µm. n = 6 sections/mouse were analyzed for n = 3 mice per condition. b) Quantification of AQP4 vessel coverage for control and N-γ1-KO mice. *P = 0.028 for 6 sections analyzed per mouse, averaged across n = 3 mice per condition, by unpaired, two-tailed t-test. c) Tilescan of one coronal section of the brain 3 days after intracardiac delivery of tdTomato-E0771 to either N-γ1-KO mice or control littermates. Scale bar = 1 mm. d) Enlarged representative images of extravasated DTCs from the tilescans in (c). Scale bar = 20 µm. e) Quantification of extravasated single tdTomato-E0771 DTCs or clusters per brain section in control and N-γ1-KO mice. P = 0.43 for n = 6 control mice and n = 5 N-γ1-KO mice, by two-tailed t-test. f) Representative bright field images of whole ovary metastases from N-γ1-KO mice or control littermates following intracardiac injection of tdTomato-E0771 cells. Ruler shown for scale. g) Quantification of volume of ovary metastases in control and N-γ1-KO mice. P = 0.066, for n = 9 control littermates and n = 7 N-γ1-KO mice, by two-tailed t test. Metastases from both ovaries were counted. h) Representative fluorescent images of the whole lung of N-γ1-KO mice or their control littermates following inoculation with tdTomato-E0771 cells, 13 days post-intracardiac injection. Tumour lesions are in white. n = 6 mice for both N-γ1-KO and control littermates groups. Data are presented as mean values + /− SEM.
Extended Data Fig. 6 Knockdown of DTC dystroglycan promotes metastatic progression of E0771 hosts; this effect is saturated in N-γ1-KO mice.
a) Representative immunoblot of E0771 cells infected with non-targeting shRNA (shScramble) or shDAG1. Blot was co-stained for α-DAG, β-DAG and Lamin A/C as a loading control. Blots representative of n = 3 biologically independent experiments. b) Integrated intensity of α-DAG and β-DAG, normalized to their respective loading control (Lamin A/C) and subsequently to shScramble band on each blot. **P = 0.002, ***P = 0.0009 for n = 3 biologically independent experiments, calculated by paired two-tailed t-test. c, d) Representative immunostains of brain sections (20-25 sections/brain) from (c) control littermates or (d) N-γ1-KO mice and, 13 days after intracardiac inoculation with either shScramble (left panels) or shDAG1 (right panels) tdTom-E0771 tumour cells. Scale bar = 5 mm. Quantification of (e) total lesion size, (f) number of lesions per section, and (g) average lesion size in these mice. P-values for n = 8 control mice per group or n = 4 N-γ1-KO mice per group calculated by two-tailed t-test. Data are presented as mean values + /− SEM.
Extended Data Fig. 7 Gating strategy to flow sort tdTomato-positive brain metastases and DTCs for single cell transcriptomic analysis.
Gating strategy for isolating tdTomato + / mVenus-p27+ micrometastases and tdTomato + / mVenus-p27− macrometastases from brains of NOD-SCID mice inoculated with tdTomato-mVenus-p27K− T4-2 cells via ultrasound-guided intracardiac injection. Dead cells were excluded based on DAPI incorporation. Live cells were further gated off PE-Texas Red (for tdTomato) and FITC-A (for mVenus-p27). Non-inoculated brain cell suspension and tdTomato- or mVenus-p27-expressing T4-2s were used to set gates.
Extended Data Fig. 8 YAP localizes to the nucleus upon DTC activation in multiple models.
a) Representative IF staining for YAP1 to assess its localization and intensity in eGFP-T4-2 DTCs at different stages of metastatic progression in the brains of NOD SCID mice, ~8 weeks after intracardiac injection of eGFP-T4-2 cells. White arrowhead denotes the YAP-low nucleus of a solitary DTC. Scale bar = 50 μm. From left to right, images are representative of n = 44, 90 and 47 independent lesions imaged across n = 4 mice. b, c) YAP1 fluorescent intensity (b) and subcellular location (c) were quantified for n = 211 total lesions across 4 mice. P-values were calculated by one-way ANOVA and Tukey’s post hoc test. d) Representative IF staining for YAP1 in metastatic lesions of N-γ1-KOs and control mice inoculated with either shScramble E0771 or shDAG1 E0771. Scale bar = 10 μm for top left panel and 100 μm for other panels. Images are representative of n = 3 sections imaged per mouse across n = 3 mice per condition. e) Co-immunoprecipitation (IP) of β-DAG and YAP1. FLAG-YAP1-transduced MDA-MB-231 cells were grown on type I Collagen (Col-1) or laminin-211 (LN-211)-coated plates. ~1 mg of protein lysate was precipitated with 40 ul of FLAG-tag monoclonal antibody or control rat IgG agarose. Precipitated proteins were blotted with YAP1 and β-DAG antibodies. ~10% of the protein lysate was used as a loading control for the input. f) Ratio of bound β-DAG that co-immunoprecipitated with YAP1 to total β-DAG in 10% of the cell lysate for MDA-MB-231 cells grown on LN-211, normalized to the same ratio from MDA-MB-231 cells grown on Col-I to correct for the variations between independent replicates. Blots are representative of n = 5 biologically independent samples probed. *P = 0.02 calculated by paired, two-tailed t-test. Data are presented as mean values + /− SEM.
Supplementary information
Supplementary Table 1
Data on patients with breast cancer associated with specimens analyzed in Fig. 4a–c.
Supplementary Table 2
Differential gene expression analysis results of Leiden-defined clusters from single-cell-profiled brain metastases and micrometastases. Data analyzed using two-sided Wilcoxon rank-sum test with Benjamini–Hochberg multiple testing correction.
Supplementary Table 3
shRNA sequences targeting human DAG1, ITGA6, ITGB1 and YAP1 and murine DAG1.
Source data
Figure 1
Statistical Source data.
Figure 2
Statistical Source data.
Figure 3
Statistical Source data.
Figure 4
Statistical Source data.
Figure 5
Statistical Source data.
Figure 5
Unprocessed immunoblots.
Figure 7
Statistical Source data.
Figure 7
Unprocessed immunoblots.
Extended Data Fig. 2
Statistical Source data.
Extended Data Fig. 3
Statistical Source data.
Extended Data Fig. 4
Statistical Source data.
Extended Data Fig. 5
Statistical Source data.
Extended Data Fig. 6
Statistical Source data.
Extended Data Fig. 6
Unprocessed immunoblots.
Extended Data Fig. 8
Statistical Source data.
Extended Data Fig. 8
Unprocessed immunoblots.
Rights and permissions
About this article
Cite this article
Dai, J., Cimino, P.J., Gouin, K.H. et al. Astrocytic laminin-211 drives disseminated breast tumor cell dormancy in brain. Nat Cancer 3, 25–42 (2022). https://doi.org/10.1038/s43018-021-00297-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-021-00297-3
This article is cited by
-
Cell-cell communication characteristics in breast cancer metastasis
Cell Communication and Signaling (2024)
-
Lung endothelium exploits susceptible tumor cell states to instruct metastatic latency
Nature Cancer (2024)
-
From pre-clinical to translational brain metastasis research: current challenges and emerging opportunities
Clinical & Experimental Metastasis (2024)
-
Harnessing immunotherapy for brain metastases: insights into tumor–brain microenvironment interactions and emerging treatment modalities
Journal of Hematology & Oncology (2023)
-
Intravital imaging to study cancer progression and metastasis
Nature Reviews Cancer (2023)