Abstract
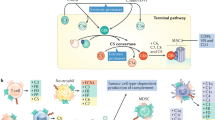
Complement has emerged as a component of tumor-promoting inflammation. We conducted a systematic assessment of the role of complement activation and effector pathways in sarcomas. C3−/−, MBL1/2−/− and C4−/− mice showed reduced susceptibility to 3-methylcholanthrene sarcomagenesis and transplanted sarcomas, whereas C1q and factor B deficiency had marginal effects. Complement 3a receptor (C3aR), but not C5aR1 and C5aR2, deficiency mirrored the phenotype of C3−/− mice. C3 and C3aR deficiency were associated with reduced accumulation and functional skewing of tumor-associated macrophages, increased T-cell activation and response to anti-PD-1 therapy. Transcriptional profiling of sarcoma-infiltrating macrophages and monocytes revealed the enrichment of the major histocompatibility complex II–dependent antigen presentation pathway in C3-deficient cells. In patients, C3aR expression correlated with a macrophage population signature, and C3-deficiency-associated signatures predicted better clinical outcome. These results suggest that the lectin pathway and C3a–C3aR axis are key components of complement and macrophage-mediated sarcoma promotion and immunosuppression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data are present in the main text and/or in the supplementary information. RNA-seq datasets are available in the NCBI Gene Expression Omnibus under accession number GSE141692. Data related to human sarcoma were downloaded from the cBioportal platform (http://www.cbioportal.org/study/summary?id=sarc_tcga). Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
All custom scripts developed to perform the analyses are available at https://figshare.com/articles/software/Custom_R_scripts_related_to_fig5_fig7_and_suppl_fig5/13286225.
References
Balkwill, F. & Mantovani, A. Inflammation and cancer: back to Virchow? Lancet 357, 539–545 (2001).
Mantovani, A., Allavena, P., Sica, A. & Balkwill, F. Cancer-related inflammation. Nature 454, 436–444 (2008).
Coussens, L. M., Zitvogel, L. & Palucka, A. K. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science 339, 286–291 (2013).
Qian, B. Z. & Pollard, J. W. Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 (2010).
Mantovani, A., Marchesi, F., Malesci, A., Laghi, L. & Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399–416 (2017).
Mantovani, A. & Allavena, P. The interaction of anticancer therapies with tumor-associated macrophages. J. Exp. Med. 212, 435–445 (2015).
Bottazzi, B., Doni, A., Garlanda, C. & Mantovani, A. An integrated view of humoral innate immunity: pentraxins as a paradigm. Annu. Rev. Immunol. 28, 157–183 (2010).
Reis, E. S., Mastellos, D. C., Ricklin, D., Mantovani, A. & Lambris, J. D. Complement in cancer: untangling an intricate relationship. Nat. Rev. Immunol. 18, 5–18 (2018).
Roumenina, L. T., Daugan, M. V., Petitprez, F., Sautes-Fridman, C. & Fridman, W. H. Context-dependent roles of complement in cancer. Nat. Rev. Cancer 19, 698–715 (2019).
Golay, J. et al. CD20 levels determine the in vitro susceptibility to rituximab and complement of B-cell chronic lymphocytic leukemia: further regulation by CD55 and CD59. Blood 98, 3383–3389 (2001).
Fishelson, Z. & Kirschfink, M. Complement C5b-9 and cancer: mechanisms of cell damage, cancer counteractions, and approaches for intervention. Front. Immunol. 10, 752 (2019).
Surace, L. et al. Complement is a central mediator of radiotherapy-induced tumor-specific immunity and clinical response. Immunity 42, 767–777 (2015).
West, E. E., Kolev, M. & Kemper, C. Complement and the regulation of T cell responses. Annu. Rev. Immunol. 36, 309–338 (2018).
Ning, C. et al. Complement activation promotes colitis-associated carcinogenesis through activating intestinal IL-1β/IL-17A axis. Mucosal Immunol. 8, 1275–1284 (2015).
Ajona, D., Ortiz-Espinosa, S. & Pio, R. Complement anaphylatoxins C3a and C5a: emerging roles in cancer progression and treatment. Semin. Cell Dev. Biol. 85, 153–163 (2019).
Afshar-Kharghan, V. The role of the complement system in cancer. J. Clin. Invest. 127, 780–789 (2017).
Markiewski, M. M. et al. Modulation of the antitumor immune response by complement. Nat. Immunol. 9, 1225–1235 (2008).
Vadrevu, S. K. et al. Complement c5a receptor facilitates cancer metastasis by altering T-cell responses in the metastatic niche. Cancer Res. 74, 3454–3465 (2014).
Cao, Q., McIsaac, S. M. & Stadnyk, A. W. Human colonic epithelial cells detect and respond to C5a via apically expressed C5aR through the ERK pathway. Am. J. Physiol. Cell Physiol. 302, C1731–C1740 (2012).
Roumenina, L. T. et al. Tumor cells hijack macrophage-produced complement C1q to promote tumor growth. Cancer Immunol. Res. 7, 1091–1105 (2019).
Bonavita, E. et al. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell 160, 700–714 (2015).
Rubino, M. et al. Epigenetic regulation of the extrinsic oncosuppressor PTX3 gene in inflammation and cancer. Oncoimmunology 6, e1333215 (2017).
Hajishengallis, G., Reis, E. S., Mastellos, D. C., Ricklin, D. & Lambris, J. D. Novel mechanisms and functions of complement. Nat. Immunol. 18, 1288–1298 (2017).
Ponzetta, A. et al. Neutrophils driving unconventional T cells mediate resistance against murine sarcomas and selected human tumors. Cell 178, 346–360 (2019).
Katenkamp, D., Kosmehl, H. & Langbein, L. Inadequate tumor surgery of chemically induced soft tissue sarcomas—an experimental approach for induction of metastasis formation? Exp. Pathol. 34, 209–215 (1988).
Li, Q. et al. Transformation potential of bone marrow stromal cells into undifferentiated high-grade pleomorphic sarcoma. J. Cancer Res. Clin. Oncol. 136, 829–838 (2010).
Molgora, M. et al. IL-1R8 is a checkpoint in NK cells regulating anti-tumour and anti-viral activity. Nature 551, 110–114 (2017).
Taylor, B. S. et al. Advances in sarcoma genomics and new therapeutic targets. Nat. Rev. Cancer 11, 541–557 (2011).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Cho, M. S. et al. Autocrine effects of tumor-derived complement. Cell Rep. 6, 1085–1095 (2014).
Nunez-Cruz, S. et al. Genetic and pharmacologic inhibition of complement impairs endothelial cell function and ablates ovarian cancer neovascularization. Neoplasia 14, 994–1004 (2012).
Guglietta, S. et al. Coagulation induced by C3aR-dependent NETosis drives protumorigenic neutrophils during small intestinal tumorigenesis. Nat. Commun. 7, 11037 (2016).
Nabizadeh, J. A. et al. The complement C3a receptor contributes to melanoma tumorigenesis by inhibiting neutrophil and CD4+ T cell responses. J. Immunol. 196, 4783–4792 (2016).
Doerner, S. K. et al. High-fat diet-induced complement activation mediates intestinal inflammation and neoplasia, independent of obesity. Mol. Cancer Res. 14, 953–965 (2016).
Wang, Y. et al. Autocrine complement inhibits IL10-dependent T-cell-mediated antitumor immunity to promote tumor progression. Cancer Discov. 6, 1022–1035 (2016).
Boire, A. et al. Complement component 3 adapts the cerebrospinal fluid for leptomeningeal metastasis. Cell 168, 1101–1113 (2017).
Ajona, D. et al. A combined PD-1/C5a blockade synergistically protects against lung cancer growth and metastasis. Cancer Discov. 7, 694–703 (2017).
de Visser, K. E., Korets, L. V. & Coussens, L. M. Early neoplastic progression is complement independent. Neoplasia 6, 768–776 (2004).
Medler, T. R. et al. Complement C5a fosters squamous carcinogenesis and limits T cell response to chemotherapy. Cancer Cell 34, 561–578 (2018).
Riihila, P. et al. Complement component C3 and complement factor B promote growth of cutaneous squamous cell carcinoma. Am. J. Pathol. 187, 1186–1197 (2017).
Aykut, B. et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574, 264–267 (2019).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Piao, C. et al. Complement 5a enhances hepatic metastases of colon cancer via monocyte chemoattractant protein-1-mediated inflammatory cell infiltration. J. Biol. Chem. 290, 10667–10676 (2015).
Czermak, B. J. et al. In vitro and in vivo dependency of chemokine generation on C5a and TNF-α. J. Immunol. 162, 2321–2325 (1999).
Venkatesha, R. T., Berla Thangam, E., Zaidi, A. K. & Ali, H. Distinct regulation of C3a-induced MCP-1/CCL2 and RANTES/CCL5 production in human mast cells by extracellular signal regulated kinase and PI3 kinase. Mol. Immunol. 42, 581–587 (2005).
Roca, H. et al. CCL2 and interleukin-6 promote survival of human CD11b+ peripheral blood mononuclear cells and induce M2-type macrophage polarization. J. Biol. Chem. 284, 34342–34354 (2009).
Sica, A. & Mantovani, A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122, 787–795 (2012).
Cho, M. S. et al. Complement component 3 is regulated by TWIST1 and mediates epithelial-mesenchymal transition. J. Immunol. 196, 1412–1418 (2016).
Petitprez, F. et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 577, 556–560 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Lai, L. et al. GSKB: a gene set database for pathway analysis in mouse. Preprint at bioRxiv https://doi.org/10.1101/082511 (2016).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128 (2013).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 14, 7 (2013).
Magrini, E. et al. Endothelial deficiency of L1 reduces tumor angiogenesis and promotes vessel normalization. J. Clin. Invest. 124, 4335–4350 (2014).
Acknowledgements
We thank F. S. Colombo for cell sorting experiments, J. Cibella for technical help in RNA-seq experiments and F. Grizzi for technical help in immunohistochemistry experiments. The contributions of Fondazione Cariplo (Contract no. 2016-0568 to E.M.); Ministero della Salute (RF-2013-02355470 to C.G.); the European Commission (ERC project PHII-669415), Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR) (PRIN 2015YYKPNN and 20174T7NXL) and Associazione Italiana Ricerca sul Cancro (AIRC IG-19014 to A.M. and AIRC IG-21714 to C.G.) and National Institutes of Health (AI068730 to J.D.L) are gratefully acknowledged. E.M. has been the recipient of a fellowship from Fondazione Umberto Veronesi (FUV, 2016–2017). We acknowledge CINECA and ELIXIR for the availability of high-performance computing resources and support.
Author information
Authors and Affiliations
Contributions
E.M. designed and conducted the experiments and drafted the manuscript. S.D.M. contributed to in vitro and in vivo experiments. C. Perucchini, M.G.L., A.P. and D.S. provided technological support for in vivo experiments. F.P., P.C. and F.C. contributed to histological analysis. C. Peano performed preparation of samples for RNA-seq analysis. S.N.M., A. Donato and R.C. analyzed RNA-seq and human TCGA data. A.I., S.J. and A. Doni contributed to the experimental design and data analysis. E.S.R. and J.D.L. provided gene-targeted animals and contributed to the experimental design and data analysis. A.M. and C.G. contributed to the experimental design and the supervision of the study and wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
All authors except for J.D.L. declare no competing interests. J.D.L. is the founder of Amyndas Pharmaceuticals, which is developing complement inhibitors for therapeutic purposes and is the inventor of patents or patent applications that describe the use of complement inhibitors for therapeutic purposes, some of which are developed by Amyndas Pharmaceuticals. J.D.L. is also the inventor of the compstatin technology licensed to Apellis Pharmaceuticals (4(1MeW)7W/POT-4/APL-1 and PEGylated derivatives such as APL-2/pegcetacoplan).
Additional information
Peer review information Nature Cancer thanks Wolf Fridman, David Kirsch and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The lectin pathway and the C3aR promote 3-MCA-induced sarcomagenesis and FS6 sarcoma growth.
a−h, 3-MCA-induced sarcoma growth curves (left panels) or mean tumor volume (± SEM, right panels) in C3−/− (a, n = 20 wt, n = 18 ko), MBL1/2−/− (b, n = 10 wt, n = 14 ko), C4−/− (c, n = 10 mice in each group), C1q−/− (d, n = 7 wt, n = 8 ko), fB−/− (e, n = 9 wt, n = 12 ko), C5aR1−/− (f, n = 10 wt, n = 19 ko), C5aR2−/− (g, n = 14 wt, n = 10 ko) and C3aR−/− (h, n = 14 wt, n = 16 ko) mice. Experiment of panel d was performed using wt littermates. i, Representative H&E staining of 3-MCA tumor tissues of wt, C3−/−, MBL1/2−/−, C4−/− and C3aR−/− mice. One experiment performed, three representative fields have been acquired for each group (n = 4 mice in each group). Scale bar: 100 µm. j-q, FS6 tumor incidences in C3−/− (j, n = 4 wt, n = 8 ko); MBL1/2−/− (k, n = 10 wt, n = 9 ko); C4-/ (l, n = 11 wt, n = 10 ko); C3aR−/− (m, n = 15 wt, n = 12 ko); C1q−/− (n, n = 15 wt, n = 8 ko); fB−/− (o, n = 9 wt, n = 4 ko); C5aR1−/− (p, n = 12 wt, n = 5 ko) and C5aR2−/− (q, n = 12 wt, n = 8 ko) mice. r-u, FS6 tumor volumes (mean ± SEM) (r, t) and incidences (s, u) in MBL1/2±/- (r-s, n = 12 he, n = 8 ko) and C3aR±/- (t-u, n = 21 he, n = 12 ko) littermates. One representative experiment out of three (a), two (b, d, e, f, j and k) or one (c, g, h, l, n, p, q, r, s, t and u) performed is shown. o, m: two pooled experiments. The same wt mice were used simultaneously as control mice of experiments of panels p, q and one out of the two pooled experiments of panel m. Two-tailed Mann Whitney test (r) or unpaired two-tailed Student’s t test (t). Exact p values are reported, two-tailed Wilcoxon matched-pairs signed rank test (j-q, s and u).
Extended Data Fig. 2 Complement deposition on sarcoma cells.
a, Flow cytometry analysis of C3b, iC3b, C3c deposition on cells derived from peritoneal washes of wt mice incubated with heat inactivated (HI) or normal serum. Representative FACS plot from a single mouse (right panel) and quantification of C3b, iC3b, C3c-positive cells on total cells is shown (left panel) (n = 6 different mice, mean ± SEM). b, Representative FACS plots showing the staining with digoxigenin-labeled DSA, GNA, MAA and SNA lectins or anti-digoxigenin alone in 3-MCA-derived sarcoma cells treated with 10 µg/ml tunicamycin (red) or vehicle (blu). c, d, Flow cytometry analysis of C3b, iC3b, C3c deposition on 3-MCA-derived (c) and MN/MCA1 (d) sarcoma cells incubated with normal serum from wt and MBL1/2−/− [n = 7 (c) and n = 5 (d) independent experiments], C1q−/− [n = 6 (c) and n = 5 (d) independent experiments], C4−/− [n = 3 (c and d) independent experiments] or fB−/− [n = 3 (c) and n = 2 (d) independent experiments] (mean ± SEM). For some experiments the same normal wt serum was used as control of different ko sera. Exact p values are reported. Two-tailed Mann Whitney test (c, C4−/− serum) or unpaired two-tailed Student’s t test (c, excluding C4−/− serum, and d).
Extended Data Fig. 3 Gating strategy for FACS analysis of TAMs and monocytes in MN/MCA1 sarcomas.
Representative FACS plots showing the gating strategy for FACS analysis of TAMs and monocytes in MN/MCA1 tumor samples. Gate of F4/80+ cells corresponds to FACS data panels of F4/80+/Aqua− (%) of Fig. 4d,j and Extended Data Fig. 4b. Gate of F4/80+CD206+ cells corresponds to FACS data panels of F4/80+CD206+/Aqua− (%) of Fig. 4f,h,k and Extended Data Fig. 4b. The expression of selected M1 markers (CD11c, MHC II, CD80 and CD86), reported as MFI in FACS data panels of Fig. 4g,i,l and Extended Data Fig. 4a,b, is gated on F4/80+ cells. Gate of Ly6C+ cells corresponds to FACS data panels of Ly6C+/Aqua- (%) of Fig. 4e,j and Extended Data Fig. 4a,b.
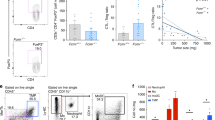
Extended Data Fig. 4 Effect of C3 and C3aR deficiency on TAMs and T cells in i.m. injected MN/MCA1 and FS6 models.
a, Analysis by FACS of M2 macrophage frequency (F4/80+CD206+) and of selected M1 marker (CD11c, MHC II, measured as MFI) expression gated on total macrophages (F4/80+) in wt and C3−/− mice sacrificed 27 days after i.m. MN/MCA1 tumor cell injection (n = 6 wt mice, n = 5 ko mice, mean ± SEM). b, Analysis by FACS of monocyte (Ly6C+), macrophage (F4/80+), M2 macrophage frequency (F4/80+CD206+) and of CD86 (measured as MFI) expression gated on total macrophages (F4/80+) in wt and C3aR−/− mice sacrificed 34 days after FS6 tumor cell injection (n = 9 wt mice, n = 4 ko mice; mean ± SEM). c, Quantitation of vessel density, vascular area and vascular coverage by pericytes in 3-MCA-derived tumors of wt and C3−/− mice (n = 7 mice in each group; mean ± SEM). d, Frequency of CD3+CD4+, CD3+CD8+ and activated effector/effector memory T cells (CD8+CD44+CD62L−) in wt and C3aR−/− mice sacrificed 34 days after FS6 tumor cell injection (n = 9 wt mice, n = 4 ko mice; mean ± SEM). a-d: one experiment performed. Exact p values are reported, unpaired two-tailed Student’s t test or two-tailed Mann Whitney test (a-d).
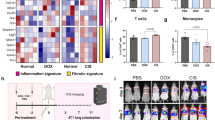
Extended Data Fig. 5 Transcriptional profiling analysis of sarcoma infiltrating monocytes.
a, Heatmap showing the first 1000 more differentially expressed genes in wt (blue) versus C3-deficient (red) monocytes. b, PC analysis of RNA expression of macrophages and monocytes. X and Y axes represent the first and the second PC, respectively. c, Number of differentially expressed mouse genes and human orthologues in tumor infiltrating leukocytes.
Extended Data Fig. 6 Gating strategy for FACS analysis of CD4+, CD8+ and CD8+CD44+CD62L− T cells in MN/MCA1 sarcomas.
Representative FACS plots showing the gating strategy for FACS analysis of CD4+, CD8+ and CD8+CD44+CD62L− T cells in MN/MCA1 tumor samples. Gate of CD4+ cells corresponds to FACS data panels of CD4+/Aqua− (%) of Fig. 6a,g and Extended Data Fig. 4d. Gate of CD8+ cells corresponds to FACS data panels of CD8+/Aqua− (%) of Fig. 6e,g and Extended Data Fig. 4d. Gate of CD8+CD44+CD62L− cells (upper right panel) corresponds to FACS data panels of CD8+CD44+CD62L−/Aqua− (%) of Fig. 6f,g and Extended Data Fig. 4d.
Extended Data Fig. 7 Complement activation and prognostic significance of C3aR expression in UPS patients.
a-d, Representative magnification images (20X) of immunostaining analysis for C1q (a), C4d (b), C3c (c) and C3aR (d) in UPS tissue sections. One experiment performed (n = 19 patients), 10 representative fields have been acquired for each patient. Representative images for patients showing negative (0% IRA, left panel) or positive (>0% IRA, central and right panels) staining for C3aR expression (d). Scale bar: 100 µm. e–f, Kaplan-Meier survival curves representing the DFS (e) and the metastasis-free survival (f) for patients showing negative (n = 5 patients) or positive (n = 14 patients) staining for C3aR expression. Exact p value of Log-rank test for survival curves, Hazard ratio (HR) and confidence intervals (CI) are indicated in the figures (e, f).
Supplementary information
Supplementary Tables
Supplementary Tables 1–11.
Source data
Source Data Fig. 1
Statistical source data for Fig. 1 and replicates.
Source Data Fig. 2
Statistical source data for Fig. 2 and replicates.
Source Data Fig. 3
Statistical source data for Fig. 3.
Source Data Fig. 4
Statistical source data for Fig. 4 and replicates.
Source Data Fig. 6
Statistical source data for Fig. 6 and replicates.
Source Data Extended Data Fig. 1
Statistical source data for Extended Data Fig. 1 and replicates.
Source Data Extended Data Fig. 2
Statistical source data for Extended Data Fig. 2.
Source Data Extended Data Fig. 4
Statistical source data for Extended Data Fig. 4.
Source Data Extended Data Fig. 7
Statistical source data for Extended Data Fig. 7.
Rights and permissions
About this article
Cite this article
Magrini, E., Di Marco, S., Mapelli, S.N. et al. Complement activation promoted by the lectin pathway mediates C3aR-dependent sarcoma progression and immunosuppression. Nat Cancer 2, 218–232 (2021). https://doi.org/10.1038/s43018-021-00173-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-021-00173-0
This article is cited by
-
A guide to complement biology, pathology and therapeutic opportunity
Nature Reviews Immunology (2024)
-
Proteomic characterization identifies clinically relevant subgroups of soft tissue sarcoma
Nature Communications (2024)
-
The proteomic landscape of soft tissue sarcomas
Nature Communications (2023)
-
Complement C3a activates astrocytes to promote medulloblastoma progression through TNF-α
Journal of Neuroinflammation (2022)
-
Macrophages as tools and targets in cancer therapy
Nature Reviews Drug Discovery (2022)