Abstract
Deep learning (DL) is a powerful methodology for the recognition and classification of tissue structures in digital pathology. Its performance in prostate cancer pathology is still under intensive investigation. Here we develop DL-based models for the detection of prostate cancer tissue in whole-slide images based on a large high-quality annotated training dataset and a modern state-of-the-art convolutional network architecture (NASNetLarge). The overall accuracy of our model for tumour detection in two validation cohorts is comparable to that of pathologists and reaches 97.3% in a native version and more than 98% using the suggested DL-based augmentation strategies. As a second step, we suggest a new biologically meaningful DL-based algorithm for Gleason grading of prostatic adenocarcinomas with high, human-level performance in prognostic stratification of patients when tested in several well-characterized validation cohorts. Furthermore, we determine the optimal minimal tumour size (real size of approximately 560 × 560 µm) for robust Gleason grading representative of the whole tumour focus. Our approach is realized in the unified digital pathology pipeline, which delivers all the relevant tumour metrics for a pathology report.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The whole-slide images used for algorithm development (training dataset) are publicly available through GDC Data Portal of the National Cancer Institute (The Cancer Genome Atlas Project; http://portal.gdc.cancer.gov). The validation datasets (image patches of tumour and benign classes) generated and analysed during the current study are available for academical use only from public repository https://zenodo.org/deposit/3825933. Any usage for publications should be consented by corresponding authors. All other data may be obtained upon request to the authors.
Code availability
The source code used in this study is available at https://github.com/gagarin37/deep_learning_pca.
References
Bera, K., Schalper, K. A., Rimm, D. L., Velcheti, V. & Madabhushi, A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 16, 703–715 (2019).
Coudray, N. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018).
Kather, J. N. et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med. 16, e1002730 (2019).
Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019).
Bychkov, D. et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 8, 3395 (2018).
Couture, H. D. et al. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. npj Breast Cancer 4, 30 (2018).
Campanella, G. et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25, 1301–1309 (2019).
Ehteshami Bejnordi, B. et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017).
Mobadersany, P. et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl Acad. Sci. USA 115, E2970–E2979 (2018).
Nagpal, K. et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. npj Digit. Med. 2, 48 (2019).
Bulten, W. et al. Epithelium segmentation using deep learning in H&E-stained prostate specimens with immunohistochemistry as reference standard. Sci. Rep. 9, 864 (2019).
Chen, C. M., Huang, Y. S., Fang, P. W., Liang, C. W. & Chang, R. F. A computer-aided diagnosis system for differentiation and delineation of malignant regions on whole-slide prostate histopathology image using spatial statistics and multidimensional DenseNet. Med. Phys. 47, 1021–1033 (2019).
Poojitha, U. P. & Lal Sharma, S. Hybrid unified deep learning network for highly precise Gleason grading of prostate cancer. In Proc. Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS 899–903 (IEEE, 2019).
Li, J. et al. A multi-scale U-Net for semantic segmentation of histological images from radical prostatectomies. AMIA Annu. Symp. Proc. 2017, 1140–1148 (2017).
Li, J. et al. An EM-based semi-supervised deep learning approach for semantic segmentation of histopathological images from radical prostatectomies. Comput. Med. Imaging Graph. 69, 125–133 (2018).
Li, W. et al. Path R-CNN for prostate cancer diagnosis and gleason grading of histological images. IEEE Trans. Med. Imaging 38, 945–954 (2019).
Lucas, M. et al. Deep learning for automatic Gleason pattern classification for grade group determination of prostate biopsies. Virchows Arch. 475, 77–83 (2019).
Ren, J. et al. Recurrence analysis on prostate cancer patients with Gleason score 7 using integrated histopathology whole-slide images and genomic data through deep neural networks. J. Med. Imaging 5, 047501 (2018).
Arvaniti, E. et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci. Rep. 8, 12054 (2018).
Singh, M. et al. Gland segmentation in prostate histopathological images. J. Med. Imaging 4, 027501 (2017).
Jiménez del Toro, O. et al. Convolutional neural networks for an automatic classification of prostate tissue slides with high-grade Gleason score. Proc. SPIE 10140, 101400O (2017).
Bulten, W. et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 21, 233–241 (2020).
Ström, P. et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 21, 222–232 (2020).
Komura, D. & Ishikawa, S. Machine learning methods for histopathological image analysis. Comput. Struct. Biotechnol. J. 16, 34–42 (2018).
Gertych, A. et al. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci. Rep. 9, 1483 (2019).
Nir, G. et al. Automatic grading of prostate cancer in digitized histopathology images: learning from multiple experts. Med. Image Anal. 50, 167–180 (2018).
Nir, G. et al. Comparison of artificial intelligence techniques to evaluate performance of a classifier for automatic grading of prostate cancer from digitized histopathologic images. JAMA Netw. Open 2, e190442 (2019).
Li, J. et al. An attention-based multi-resolution model for prostate whole slide imageclassification and localization. Preprint at https://arxiv.org/abs/1905.13208 (2019).
Gertych, A. et al. Machine learning approaches to analyze histological images of tissues from radical prostatectomies. Comput. Med. Imaging Graph. 46 (Pt 2), 197–208 (2015).
Litjens, G. et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 6, 26286 (2016).
Zoph, B., Vasudevan, V., Shlens, J. & Le, Q. V. Learning transferable architectures for scalable image recognition. Preprinit at https://arxiv.org/abs/1707.07012 (2017).
Tolkach, Y., Thomann, S. & Kristiansen, G. Three-dimensional reconstruction of prostate cancer architecture with serial immunohistochemical sections: hallmarks of tumour growth, tumour compartmentalisation, and implications for grading and heterogeneity. Histopathology 72, 1051–1059 (2018).
van Royen, M. E. et al. Three-dimensional microscopic analysis of clinical prostate specimens. Histopathology 69, 985–992 (2016).
Macenko, M. et al. A method for normalizing histology slides for quantitative analysis. In IEEE International Symposium on Biomedical Imaging: From Nano to Macro 1107–1110 (IEEE, 2009).
Tolkach, Y. & Kristiansen, G. Cribriform and glomeruloid acinar adenocarcinoma of the prostate—an intratumoural intraductal carcinoma? Histopathology 74, 804–808 (2019).
Humphrey, P. A., Moch, H., Cubilla, A. L., Ulbright, T. M. & Reuter, V. E. The 2016 WHO classification of tumours of the urinary System and male genital organs—part B: prostate and bladder tumours. Eur. Urol. 70, 106–119 (2016).
Acknowledgements
The GPU card for this study was donated by the NVIDIA corporation (a GPU academic grant programme). No funding was received for this study.
Author information
Authors and Affiliations
Contributions
Conceiving the study and the design: Y.T. Annotations, creating the datasets, developing and training the models, development of the grading algorithm and augmentation strategies, digital pathology pipeline, validation tests, hardware, conducting the statistical analysis and data analysis: Y.T. Providing validation datasets: G.K. Grading for validation experiments: Y.T., G.K. and M.T. Data interpretation: Y.T. and G.K. Drafting the manuscript: Y.T. Critically revising for important intellectual content: Y.T., T.D., M.T. and G.K.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
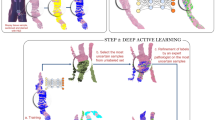
Extended Data Fig. 1 Summary of study design and setup.
Study design and setup of single study steps, principles of patch generation (including number of patches generated in training and validation datasets in thousands of unique non-intersecting patches, k), approaches to training and validation. For details to certain steps see Methods.
Extended Data Fig. 2 Training principles of the model.
Three classes were used for training (benign glandular, benign non-glandular, tumour) with stain normalization. NASNetLarge architecture was used with addition of three new layers for classification (flatten, fully connected, and classification layers). Probability of being benign could be summarized from output probabilities of two benign classes. Training principles and parameters are presented in boxes.
Extended Data Fig. 3 Deep learning – based prostate cancer grading.
a. Prostate cancer (PCA) architecture is not just a rough mix of glands with Gleason patterns (GP) 3, 4, and 5. In three dimensions, carcinoma is a tree, where single GPs do not exist, being a continuum. b. Our Gleason grading algorithm is based on the understanding of GPs being a continuum. During processing, a tumour region is being cut into patches that are then separately analyzed by convnet (trained on pure GPs, see Materials and Methods). Probabilities of different classes (GP3, GP4, GP5) are counted for every single patch and summarized for the whole tumour region and not used for nomination of every patch to a discrete GP (as in classical paradigm).
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and methods.
Rights and permissions
About this article
Cite this article
Tolkach, Y., Dohmgörgen, T., Toma, M. et al. High-accuracy prostate cancer pathology using deep learning. Nat Mach Intell 2, 411–418 (2020). https://doi.org/10.1038/s42256-020-0200-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-020-0200-7
This article is cited by
-
Künstliche Intelligenz in der Pathologie: Status quo und Zukunftsperspektiven
Die Onkologie (2024)
-
A review and comparative study of cancer detection using machine learning: SBERT and SimCSE application
BMC Bioinformatics (2023)
-
High accuracy epidermal growth factor receptor mutation prediction via histopathological deep learning
BMC Pulmonary Medicine (2023)
-
Diagnosis of autism spectrum disorder based on functional brain networks and machine learning
Scientific Reports (2023)
-
An international multi-institutional validation study of the algorithm for prostate cancer detection and Gleason grading
npj Precision Oncology (2023)