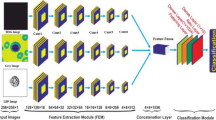
Abstract
Reliable recognition of malignant white blood cells is a key step in the diagnosis of haematologic malignancies such as acute myeloid leukaemia. Microscopic morphological examination of blood cells is usually performed by trained human examiners, making the process tedious, time-consuming and hard to standardize. Here, we compile an annotated image dataset of over 18,000 white blood cells, use it to train a convolutional neural network for leukocyte classification and evaluate the network’s performance by comparing to inter- and intra-expert variability. The network classifies the most important cell types with high accuracy. It also allows us to decide two clinically relevant questions with human-level performance: (1) if a given cell has blast character and (2) if it belongs to the cell types normally present in non-pathological blood smears. Our approach holds the potential to be used as a classification aid for examining much larger numbers of cells in a smear than can usually be done by a human expert. This will allow clinicians to recognize malignant cell populations with lower prevalence at an earlier stage of the disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The full single-cell image dataset and corresponding annotations are publicly available at The Cancer Imaging Archive (TCIA): https://doi.org/10.7937/tcia.2019.36f5o9ld 43.
Code availability
Code for the network trained in this study and network weights for one fold are available on CodeOcean, together with a subset of the single-cell image data used to test the network: https://codeocean.com/capsule/9068249/tree/v1 44.
References
Bain, B. J. Diagnosis from the blood smear. N. Engl. J. Med.353, 498–507 (2005).
Tkachuk, D. C. & Hirschmann, J. V. Wintrobe’s Atlas of Clinical Hematology (Lippincott Raven, 2006).
Theml, H., Diem, H. & Haferlach, T. Color Atlas of Hematology (Thieme, 2004).
Döhner, H. et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood129, 424–447 (2017).
Swerdlow, S. H. et al. (eds) WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues 4th edn (International Agency for Research on Cancer, 2017).
Arber, D. A. et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood127, 2391–2405 (2016).
Bennett, J. M. et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French–American–British Cooperative Group. Ann. Intern. Med.103, 620–625 (1985).
Font, P. et al. Inter-observer variance with the diagnosis of myelodysplastic syndromes (MDS) following the 2008 WHO classification. Ann. Hematol.92, 19–24 (2013).
Font, P. et al. Interobserver variance in myelodysplastic syndromes with less than 5% bone marrow blasts: unilineage vs. multilineage dysplasia and reproducibility of the threshold of 2% blasts. Ann. Hematol.94, 565–573 (2015).
Fuentes-Arderiu, X. & Dot-Bach, D. Measurement uncertainty in manual differential leukocyte counting. Clin. Chem. Lab. Med.47, 112–115 (2009).
Goodfellow, I., Bengio, Y. & Courville, A. Deep Learning (MIT Press, 2016).
Rawat, W. & Wang, Z. Deep convolutional neural networks for image classification: a comprehensive review. Neural Comput.29, 2352–2449 (2017).
Russakovsky, O. et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vision115, 211–252 (2015).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature542, 115–118 (2017).
Eulenberg, P. et al. Reconstructing cell cycle and disease progression using deep learning. Nat. Commun.8, 463 (2017).
Janowczyk, A. & Madabhushi, A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J. Pathol. Inform.7, 29 (2016).
Fuchs, T. J. & Buhmann, J. M. Computational pathology: challenges and promises for tissue analysis. Comput. Med. Imaging Graph.35, 515–530 (2011).
Albarqouni, S. et al. AggNet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans. Med. Imaging35, 1313–1321 (2016).
Levenson, R. M., Fornari, A. & Loda, M. Multispectral imaging and pathology: seeing and doing more. Expert Opin. Med. Diagn.2, 1067–1081 (2008).
Gertych, A. et al. Machine learning approaches to analyze histological images of tissues from radical prostatectomies. Comput. Med. Imaging Graph.46, 197–208 (2015).
Bigorra, L., Merino, A., Alférez, S. & Rodellar, J. Feature analysis and automatic identification of leukemic lineage blast cells and reactive lymphoid cells from peripheral blood cell images. J. Clin. Lab. Anal.31, e22024 (2017).
Krappe, S., Wittenberg, T., Haferlach, T. & Munzenmayer, C. Automated morphological analysis of bone marrow cells in microscopic images for diagnosis of leukemia: nucleus–plasma separation and cell classification using a hierarchical tree model of hematopoesis. Proc. SPIE9785, 97853C (2016).
Scotti, F. Automatic morphological analysis for acute leukemia identification in peripheral blood microscope images. In Computational Intelligence for Measurement Systems and Applications (CIMSA) 96–101 (IEEE, 2005).
Mohapatra, S., Patra, D. & Satpathy, S. An ensemble classifier system for early diagnosis of acute lymphoblastic leukemia in blood microscopic images. Neural Comput. Appl.24, 1887–1904 (2014).
Greenspan, H., van Ginneken, B. & Summers, R. M. Deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging35, 1153–1159 (2016).
Shen, D., Wu, G. & Suk, H. Deep learning in medical image analysis. Ann. Rev. Biomed. Eng.19, 221–248 (2017).
Choi, J. W. et al. White blood cell differential count of maturation stages in bone marrow smear using dual-stage convolutional neural networks. PLoS One12, e0189259 (2017).
Kainz, P., Burgsteiner, H., Asslaber, M. & Ahammer, H. Training echo state networks for rotation-invariant bone marrow cell classification. Neural Comput. Appl.28, 1277–1292 (2017).
Su, M.-C., Cheng, C.-Y. & Wang, P.-C. A neural-network-based approach to white blood cell classification. Sci. World J.2014, 796371 (2014).
Macawile, M. J., Quiñones, V. V., Ballado, A., Cruz, J. D. & Caya, M. V. White blood cell classification and counting using convolutional neural network. In 2018 3rd International Conference on Control and Robotics Engineering (ICCRE) 259–263 (IEEE, 2018).
Keohane, E. M., Smith, L. & Walenga, J. M. Rodak’s Hematology—Clinical Principles and Applications 5th edn (Elsevier, 2016).
Xie, S., Girshick, R., Dollár, P., Tu, Z. & He, K. Aggregated residual transformations for deep neural networks. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 5987–5995 (IEEE, 2017).
Dietz, M. ResNeXt implementation for Keras. GitHub Gist https://gist.githubusercontent.com/mjdietzx/ (2017).
Chollet, F. et al. Keras 2.0. Keras https://keras.io (2017).
Bychkov, D. et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep.8, 3395 (2018).
Simonyan, K., Vedaldi, A. & Zisserman, A. Deep inside convolutional networks: visualising image classification models and saliency maps. Preprint at https://arxiv.org/abs/1312.6034 (2013).
Mandrekar, J. N. Receiver operating characteristic curve in diagnostic test assessment. J. Thorac. Oncol.5, 1315–1316 (2010).
Hosmer, D. & Lemeshow, S. Applied Logistic Regression 2nd edn (Wiley, 2000).
Xing, F. & Yang, L. Robust nucleus/cell detection and segmentation in digital pathology and microscopy images: a comprehensive review. IEEE Rev. Biomed. Eng.9, 234–263 (2016).
Cuevas, E. et al. White blood cell segmentation by circle detection using electromagnetism-like optimization. Comput. Math. Methods Med.2013, 395071 (2013).
Alomari, Y. M., Abdullah, S. N. H. S., Azma, R. Z. & Omar, K. Automatic detection and quantification of WBCs and RBCs using iterative structured circle detection algorithm. Comput. Math. Methods Med.2014, 979302 (2014).
He, K., Gkioxari, G., Dollár, P. & Girshick, R. Mask R-CNN. In Proceedings of the International Conference on Computer Vision (ICCV) 2980–2988 (IEEE, 2017).
Matek, C., Schwarz, S., Spiekermann, K. & Marr, C. A single-cell morphological dataset of leukocytes from AML patients and non-malignant controls (AML-Cytomorphology_LMU). TCAI https://doi.org/10.7937/tcia.2019.36f5o9ld (2019).
Matek, C., Schwarz, S., Spiekermann, K. & Marr, C. A neural network for classifying leukocyte images from blood smears. CodeOcean https://codeocean.com/capsule/9068249/tree/v1 (2019).
Acknowledgements
We thank N. Chlis for comments on the manuscript, K. Metzeler for helpful discussions and A. Holzäpfel for contributions to the annotation task. This work was supported by the German Research Foundation DFG within the Collaborative Research Center SFB 1243. C. Matek acknowledges support from Deutsche José Carreras-Leukämie Stiftung.
Author information
Authors and Affiliations
Contributions
C. Matek, C. Marr and K.S. conceived the initial idea. C. Matek selected the cohort, digitized blood smears, wrote annotation software, and trained and evaluated the network. S.S. contributed to selecting the cohort and annotated the image data. C. Matek, C. Marr and K.S. interpreted data and wrote the paper. All authors approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Matek, C., Schwarz, S., Spiekermann, K. et al. Human-level recognition of blast cells in acute myeloid leukaemia with convolutional neural networks. Nat Mach Intell 1, 538–544 (2019). https://doi.org/10.1038/s42256-019-0101-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-019-0101-9
This article is cited by
-
CDC-NET: a cell detection and confirmation network of bone marrow aspirate images for the aided diagnosis of AML
Medical & Biological Engineering & Computing (2024)
-
A lightweight deep learning model for acute myeloid leukemia-related blast cell identification
The Journal of Supercomputing (2024)
-
Automatic identification of medically important mosquitoes using embedded learning approach-based image-retrieval system
Scientific Reports (2023)
-
Detection of acute promyelocytic leukemia in peripheral blood and bone marrow with annotation-free deep learning
Scientific Reports (2023)
-
Artificial intelligence for digital and computational pathology
Nature Reviews Bioengineering (2023)