Abstract
Prolactin (PRL) is elevated in B-cell-mediated lymphoproliferative diseases and promotes B-cell survival. Whether PRL or PRL receptors drive the evolution of B-cell malignancies is unknown. We measure changes in B cells after knocking down the pro-proliferative, anti-apoptotic long isoform of the PRL receptor (LFPRLR) in vivo in systemic lupus erythematosus (SLE)- and B-cell lymphoma-prone mouse models, and the long plus intermediate isoforms (LF/IFPRLR) in human B-cell malignancies. To knockdown LF/IFPRLRs without suppressing expression of the counteractive short PRLR isoforms (SFPRLRs), we employ splice-modulating DNA oligomers. In SLE-prone mice, LFPRLR knockdown reduces numbers and proliferation of pathogenic B-cell subsets and lowers the risk of B-cell transformation by downregulating expression of activation-induced cytidine deaminase. LFPRLR knockdown in lymphoma-prone mice reduces B-cell numbers and their expression of BCL2 and TCL1. In overt human B-cell malignancies, LF/IFPRLR knockdown reduces B-cell viability and their MYC and BCL2 expression. Unlike normal B cells, human B-cell malignancies secrete autocrine PRL and often express no SFPRLRs. Neutralization of secreted PRL reduces the viability of B-cell malignancies. Knockdown of LF/IFPRLR reduces the growth of human B-cell malignancies in vitro and in vivo. Thus, LF/IFPRLR knockdown is a highly specific approach to block the evolution of B-cell neoplasms.
Similar content being viewed by others
Introduction
B-cell malignancies, such as diffuse large B-cell lymphoma (DLBCL) and acute lymphoblastic leukemia (ALL), have widely divergent outcomes1,2. To identify early interventions for high-risk B-cell malignancies, we are delineating the mechanisms underlying the evolution of these neoplasms, which include malignancy initiation, overt neoplasm establishment, and maintenance of overt malignancy.
Because the initiation of B-cell malignancies is >3-fold more frequent in patients with the B cell-mediated autoimmune disease, systemic lupus erythematosus (SLE), than the healthy population3,4,5, SLE-prone mouse models represent robust biological systems to identify signaling pathways that elevate the risk of B-cell transformation6. Similarly, because B-cell cancers often clonally evolve from pre-malignant and indolent B-cell clones7,8,9, transgenic mice prone to developing B-cell neoplasms are robust models for investigating the mechanisms that promote the establishment of overt B-cell malignancies. For analysis of sustenance of overt B-cell malignancies, we have used cell lines and samples derived from patients with these neoplasms.
Prolactin (PRL), a sex hormone, classically recognized for its crucial role in lactation but produced by both sexes in response to acute and chronic stress10, can drive pathological processes including tumorigenesis11,12. We postulated that PRL promotes the evolution of B-cell malignancies because it (1) promotes the survival of autoreactive B cells13 and is associated with the exacerbation of SLE14,15, a disease that increases the risk of B-cell malignancy initiation3,4,5, (2) enhances the survival of normal and pathogenic mouse B cells and induces their expression of the proto-oncogenes c-MYC (cellular Myelocytomatosis) and BCL2 (B-cell lymphoma 2) in vitro16,17,18,19, and (3) complexes with the immunoglobulin G (IgG) heavy chain to promote proliferation of malignant B cells in chronic lymphocytic leukemia (CLL), a malignancy associated with autoimmune manifestations20.
Murine and human PRL receptors (PRLRs) are cytokine receptors with long (LF) and short (SF) isoforms, generated by alternative splicing21,22. Humans have an additional intermediate splice isoform (IF), at least in pathological tissues23. These different PRLRs have identical extracellular and transmembrane domains but differ in their intracellular domains such that they initiate different signals. A major signaling difference between the two categories of receptor is that the LFPRLR can activate the Janus kinase/signal transducer and activation of transcription (Jak/STAT) pathway, whereas the shorter cytoplasmic domains of the SFPRLRs cannot engage STAT11. Although the IF cannot engage STATs, it initiates STAT-independent pro-tumorigenic signaling upon dimerization with LF23,24. Overall, the increased expression of LF/IF relative to SFPRLRs on cells causes cell proliferation and survival, whereas increased expression of SF relative to LF/IF inhibits proliferation and induces apoptosis23,25,26. Thus, LF/IFPRLRs are antagonistic to SFPRLRs.
Whether PRL drives the evolution of overt B-cell malignancies via specific PRLR isoforms, is unknown. We hypothesized that, PRL enhances the risk of clonal evolution of autoimmune and pre-malignant B cells and sustains overt B-cell malignancies by signaling through the pro-proliferative and anti-apoptotic LF/IFPRLR. To test this, we measured the effects of systemically knocking down expression of the LFPRLR on immune cells: (1) in vivo in mouse models prone to SLE carrying non-malignant but aberrant B cells27, (2) in vivo in mouse models prone to DLBCL having pre-malignant B cells28, and (3) in vitro and in vivo in overt malignant human B-cell lines along with knockdown of IFPRLR.
For isoform-specific knockdown of LFPRLR in mice and LF/IFPRLR in human cells, we employed an innovative and tractable approach involving 25mer splice-modulating deoxyribonucleic acid morpholino oligomers (SMOs), termed here ‘LFPRLR SMO’, that selectively prevent splicing to produce the LFPRLR in mice and LF and IF PRLRs in humans. More specifically, the SMOs prevent the inclusion of exon 10 during splicing of PRLR pre-messenger ribonucleic acid (pre-mRNA) that encodes an intracellular domain in the LFPRLR required for binding of STAT proteins. The sequence of LFPRLR SMO is different for mouse and human29. The SMOs are highly stable, non-immunogenic deoxyribonucleic acid (DNA) oligomers linked to an octaguanidine dendrimer that ensures effective whole-body uptake. They are administered subcutaneously and have been found to be efficacious and non-toxic in healthy mice and syngeneic and human xenograft models of breast cancer29,30.
We found that signaling of PRL through the LF/IFPRLR raises the risk of B-cell malignancy initiation in SLE-prone mice, increases the survival of pre-malignant B cells in DLBCL-prone mice, and sustains the progression of established human B-cell malignancies. High expression of PRL in tumors associates with poor clinical prognosis in patients with B-cell malignancies. We identify blocking synthesis of the LF/IFPRLR using LFPRLR SMO as an isoform-specific and clinically attractive strategy that merits further investigation as a potential treatment for autoimmune and malignant B cell-mediated lymphoproliferative diseases.
Results
Knockdown of LFPRLR reduces splenic B-cell subsets in SLE-prone mice
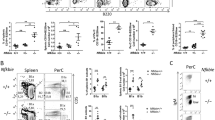
To investigate whether the PRL-LFPRLR axis raises the risk of initiation of B-cell malignancies, we compared SLE-prone MRL-lpr mice treated with either control SMO or LFPRLR SMO. Among SLE-prone models, we chose MRL-lpr mice because they accumulate genetic lesions indicative of B-cell transformation6 and exhibit the spontaneous lymphoproliferation mutation Faslpr27 that has been implicated in lymphoma pathogenesis31,32. Female MRL-lpr mice succumb to SLE between 17–22 weeks postnatally27. Therefore, we treated female mice with control SMO or LFPRLR SMO from the age of 6 weeks until 14 weeks when we quantified expression of molecular indicators known to raise the risk of B-cell malignancy development (Fig. 1a).
a–c Experimental design (a), verification of LFPRLR knockdown in total splenic WBCs by qPCR (b), and splenic WBC counts (c) in MRL-lpr mice treated with control SMO [n = 9 for (b) and n = 14 for (c), red] or LFPRLR SMO [n = 10 for (b) and n = 14 for (c), blue]. d–f Quantitation and representative flow cytometry plots of CD3−CD19+ B cells and CD19−CD3+ T cells (d); CD11c+PDCA1+ pDCs and CD11c+PDCA1− cDCs (e); Blimp1+CD138+ total plasma cells, Blimp1+CD138+B220+ plasmablasts, and Blimp1+CD138+B220− long lived plasma cells (f) in spleens of MRL-lpr mice treated with control SMO (n = 14, red) or LFPRLR SMO (n = 12, blue). Graphs show median ± interquartile range. Each dot in qPCR analyses of transcripts corresponds to the mean expression of that gene in one mouse calculated from 3 technical replicates. Ubiquitin B (Ubb) was used as the house-keeping gene in qPCR. Exact p-values were calculated using the Mann–Whitney U test. ns non-significant.
Because no commercially available antibodies could detect endogenous expression of PRLR isoforms in our models, we used qPCR to measure the absolute levels of each isoform. Of the mouse Prlr isoforms, LF, SF1, SF2, and SF322, only SF3 and LF were found to be expressed in splenic leukocytes of MRL-lpr mice. The ratio of LF Prlr: total Prlr mRNA, which indicates the level of pro-proliferative and/or anti-apoptotic PRLR signaling in cells, was significantly downregulated in splenic white blood cells (WBCs) of MRL-lpr mice treated with LFPRLR SMO (Fig. 1b). This confirmed LFPRLR knockdown in immune cells.
Because splenic T-, B-, and dendritic cells (DCs) drive SLE pathology33, we examined changes in these WBCs after LFPRLR knockdown by flow cytometry (Supplementary Fig. 1). WBC, B-cell, plasmacytoid (pDC), and conventional DC (cDC) counts were significantly reduced after LFPRLR knockdown, whereas counts of T cells and T-cell subsets (CD4+, CD8+, and pathologic CD4−CD8− SLE T cells) remained unaltered (Fig. 1c–e, Supplementary Fig. 2).
B-cell pathology in SLE is largely driven by autoantibody-producing Blimp1+CD138+B220+ short-lived plasmablasts and Blimp1+CD138+B220− long-lived plasma cells (LLPCs)34. B cells mature into plasma cells with help from DCs35,36,37,38,39,40,41. Reductions in B cells and DCs after LFPRLR knockdown suggested that treatment with LFPRLR SMO may affect plasma cell subsets. As predicted, plasmablast and LLPC numbers were significantly reduced after LFPRLR knockdown (Fig. 1f, Supplementary Fig. 3a).
As SLE progresses, there is an increase in production of autoantibodies such as those against double-stranded (ds) DNA42. Furthermore, a recent study showed that PRL drives the production of anti-dsDNA autoantibodies in MRL-lpr mice43. Whether PRL promotes such autoantibody production by signaling specifically through the LFPRLR was not determined. Hence, we determined whether LFPRLR knockdown impacted the production of anti-dsDNA autoantibodies in serum of SLE-prone mice. Although the early stage of disease analyzed makes the Mrl-lpr model less than ideal for this measurement, levels of anti-dsDNA autoantibodies trended towards reduction after LFPRLR knockdown (Supplementary Fig. 4), which matches with the reduction in plasma cell subsets seen earlier. Our findings lead us to conclude that LFPRLR knockdown reduces numbers of pathologic splenic B-cell subsets in SLE-prone mice.
Knockdown of LFPRLR in SLE-prone mice decreases factors associated with the risk of lymphoma initiation
We examined B-cell turnover and composition of the mature B-cell repertoire in SLE-prone mice post LFPRLR knockdown. We confirmed that treatment with LFPRLR SMO reduces the LF Prlr: total Prlr ratio in magnetically sorted splenic B cells (Supplementary Fig. 5) by qPCR (Fig. 2a). We then measured B-cell proliferation and found that LFPRLR knockdown reduced the percentage of B cells in S phase while concomitantly increasing the proportion of G0/G1 B cells (Fig. 2b). LFPRLR knockdown did not increase the frequency of pre-apoptotic sub-G0/G1 B cells, concordant with Fas dysfunctionality in MRL-lpr mice that prevents the death of arrested B cells.
a–d Verification of LFPRLR knockdown in isolated B cells by qPCR (a), percentages of B cells in sub G0/G1, G0/G1, S, and G2/M phases with representative flow cytometry plots (b), B-cell specific transcript levels of Myc, Bcl2 by qPCR (c), and BCL2 protein by flow cytometry (d) in spleens of MRL-lpr mice treated with control SMO (n = 14, red) or LFPRLR SMO (n = 14, blue). e–g Next-generation sequencing of total IGH repertoire (GSE207186) to compare spectratype of splenic B cells (e), and frequencies of splenic B cells with IGH CDR3 ≥ 20aa (f), and frequencies of B cells with non-productive IGH rearrangements (g) in MRL-lpr SLE-prone mice treated with control (n = 6, red) or LFPRLR SMO (n = 6, blue). h Aicda mRNA and protein levels in splenic B cells and representative histograms for AID protein staining in control SMO-treated (n = 14, red) and LFPRLR SMO-treated (n = 14, blue) MRL-lpr mice by flow cytometry. Splenic WBCs from wildtype mice cultured with IL-4 and LPS were used as positive controls for AID staining. Each dot in qPCR analyses of transcripts corresponds to the mean expression of that gene in one mouse calculated from 3 technical replicates. Ubb was used as the house-keeping gene in qPCR. Graphs show median ± interquartile range. Exact p-values were calculated using the Mann–Whitney U test. ns non-significant, FMO fluorescence minus one control, MFI median fluorescence intensity.
Because PRL induces the expression of MYC and BCL2 in lymphocytes16,17,18,19, we examined the expression of these proto-oncogenes in splenic B cells after LFPRLR knockdown. We found no changes in Myc, significantly reduced Bcl2 mRNA, but no changes in BCL2 protein in the time frame of treatment (Fig. 2c, d, Supplementary Fig. 3b). The anti-apoptotic protein BCL2 is known to promote B-cell survival but not proliferation44. Our findings showing unchanged expression of BCL2 protein and reduction in B-cell proliferation after LFPRLR knockdown in SLE-prone mice are consistent with this. However, the significant reduction in Bcl2 mRNA suggests the possibility of longer-term downregulation of BCL2 protein by LFPRLR knockdown.
Next, we investigated whether LFPRLR induced indicators of evolution of autoimmune B cells into malignancies by comparing the splenic B-cell repertoire of SLE-prone mice treated with control SMO (n = 6) or LFPRLR SMO (n = 6) using immunoglobulin heavy chain (IGH) sequencing. A healthy B-cell repertoire has very few deleterious, potentially polyreactive B-cell clones with IGH complementary determining region 3 (CDR3) of lengths ≥20 amino acids (≥60 nucleotides)45,46,47,48 and B cells carrying non-functional IGH rearrangements with an increased propensity towards malignant transformation9. Although the B-cell repertoire was normally distributed after LFPRLR knockdown, frequencies of potentially abnormal B cells with long CDR3s ranging from 35-42 amino acids, and those with non-functional IGH rearrangements were significantly reduced in LFPRLR SMO-treated SLE-prone mice (Fig. 2e–g).
B-cell malignancies evolve from hyperactivation of, and off-target genomic alterations induced by, the B-cell enzyme, activation induced cytidine deaminase (AID, Aicda gene)9,49. AID diversifies the B-cell repertoire through somatic hypermutation (SHM) and class switch recombination (CSR)50. We hypothesized that PRL-LFPRLR signaling may induce AID expression because (1) estradiol, which promotes Aicda transcription and accelerates malignant transformation of B-lymphocytes in MRL-lpr mice51 could, in part, mediate this process via stimulation of PRL secretion, and (2) a reduction in both DCs that mediate CSR, and LLPC that result from class-switched B cells in LFPRLR SMO-treated SLE-prone mice suggested that PRL-LFPRLR signaling may regulate AID expression. As anticipated, expression of Aicda mRNA and AID protein was significantly reduced in splenic B cells after LFPRLR knockdown (Fig. 2h, Supplementary Fig. 3c). Hence, PRL signaling through LFPRLR primes B cells for acquisition of oncogenic mutations by promoting AID expression. Thus, the PRL-LFPRLR axis promotes the retention of deleterious B cells, thereby increasing the pool of B cells available for malignant transformation and malignancy initiation.
Knockdown of LFPRLR impacts early B-lymphopoiesis in SLE-prone mice
We determined the extent to which reduced splenic B cells after LFPRLR knockdown in SLE-prone mice resulted from changes in early B-lymphopoiesis. Although bone marrow B-cell counts were unchanged after LFPRLR knockdown, their expression of BCL2 was strikingly reduced (Fig. 3a, b). Because BCL2 is critical for survival of immature B cells52, including autoreactive B cells46,53,54, we conclude that the LFPRLR likely contributes to the inappropriate survival and retention of abnormal, early B cells in SLE-prone mice.
a Quantitation and representative flow cytometry plots of bone marrow CD19+ B cells in MRL-lpr mice treated with control SMO (n = 9, red) or LFPRLR SMO (n = 9, blue). b Quantitation and representative histograms showing median fluorescence intensity (MFI) of BCL2 protein in bone marrow B cells of control SMO-treated (n = 5, red) and LFPRLR SMO-treated (n = 5, blue) MRL-lpr mice by flow cytometry. c A model depicting the mechanisms by which LFPRLR increases the number of abnormal splenic B cells available for potential malignant transformation or the concurrent worsening of autoimmune pathology in SLE-prone mice. Graphs show median ± interquartile range. Exact p-values were calculated using the Mann–Whitney U test. ns non-significant, FMO fluorescence minus one control. c was created with BioRender.com.
Overall, LFPRLR drives inappropriate early and mature B-lymphopoiesis in SLE-prone mice; the former mediated via induction of BCL2 expression and the latter by increasing DC numbers, which can in turn promote mature B-cell cycling, AID expression, and plasma cell generation. Consequently, deleterious B cells with non-productive IGH rearrangements and long CDR3s are retained in the repertoire, thereby raising the risk of lymphomagenesis (Fig. 3c).
Knockdown of LFPRLR in DLBCL-prone mice suppresses factors that can drive overt B-cell lymphomagenesis
To determine whether the PRL-LFPRLR axis elevates the risk of establishment of overt B-cell malignancies from pre-malignant B cells, we compared B-cell pathology in DLBCL-prone CD79b-TCL1-tg mice28, treated with either control SMO or LFPRLR SMO. All mice hemizygous for the T-cell leukemia/lymphoma protein 1 (TCL1) transgene develop signs of increased risk of DLBCL beginning at 4 months of age. These include mild lymphocytosis and slight increases (<2-fold) in WBC counts, without changes in lymphoid organ size or structure, or changes in early or late B-cell lymphopoiesis. However, the majority (~95%) of mice become visibly ill with symptoms of overt DLBCL between 7–12 months of age28. Therefore, to examine the role of LFPRLR in raising the risk of development of overt lymphoma from pre-malignant B cells, we treated 8-week-old TCL1-tg mice with the SMOs until 16-weeks-old (Fig. 4a), which, in every mouse, is before the development of overt DLBCL. As in MRL-lpr SLE-prone mice, only SF3 and LF were found to be expressed in splenic leukocytes of TCL1-tg DLBCL-prone mice. We first confirmed LFPRLR knockdown in total splenic WBCs by qPCR (Fig. 4b). LFPRLR knockdown significantly reduced the numbers of splenic WBC in TCL1-tg DLBCL-prone mice (Fig. 4c). Among splenic WBC subsets, LFPRLR knockdown significantly reduced B-cell counts but not counts of total T cells, T-cell subsets, or DC subsets (Fig. 4d, Supplementary Fig. 6). LFPRLR knockdown also significantly reduced expression of the driver TCL1 oncoprotein28 in splenic B cells in the DLBCL-prone mice (Fig. 4e, Supplementary Fig. 3d). Hence, abrogation of LFPRLR synthesis in 16-week-old TCL1-tg-mice specifically impacts pre-malignant B cells and reduces the major indicators of B-cell lymphoma risk specific to this age group. These findings strengthened the rationale for further interrogating the causal mechanisms by which LFPRLR raises the risk for establishment of overt B-cell neoplasms.
a–c Experimental design (a), verification of LFPRLR knockdown in total splenic WBCs by qPCR (b), and splenic WBC counts (c) in TCL1-tg DLBCL-prone mice treated with control SMO [n = 8 for (b) and n = 16 for (c), red] or LFPRLR SMO [n = 8 for (b) and n = 14 for (c), blue]. d–i Quantitation and representative flow cytometry plots of CD3−CD19+ B cells (d, h), splenic B cell-specific expression of TCL1 (e), and B cell-specific expression of BCL2 (f, g, i) in spleen and bone marrow of TCL1-tg mice treated with control SMO [n = 14 for (d), n = 6 for (e), n = 4 for (f), n = 8 for (g) and n = 10 for (h, i), red] or LFPRLR SMO [n = 12 for (d), n = 4 for (e), n = 4 for (f), n = 8 for (g) and n = 9 for (h, i), blue]. One representative histogram from each group for measurement of TCL1 protein in spleen and BCL2 protein in spleen and bone marrow is shown. j PRL-LFPRLR signaling upregulates BCL2 and increases survival of pre-malignant B cells, thereby enhancing the risk of establishment of overt B-cell malignancies. Graphs show median ± interquartile range. Each dot in qPCR analyses of transcripts corresponds to the mean expression of that gene in pooled samples from 3 mice that were run in 3 technical replicates. Ubb was used as the house keeping gene in qPCR. Exact p-values were calculated using the Mann–Whitney U test. ns non-significant, FMO fluorescence minus one control, MFI median fluorescence intensity. j was created with BioRender.com.
B-cell cycling was unaffected after LFPRLR knockdown in TCL1-tg mice (Supplementary Fig. 7). To examine whether LFPRLR knockdown modulated expression of Myc and Bcl2 mRNA, we initially used splenic WBCs because we could sort only a limited number of B cells even after pooling samples from ~3–4 mice per group. Myc expression was unchanged but that of Bcl2 mRNA was significantly downregulated after LFPRLR knockdown (Supplementary Fig. 8). With the few sorted pooled splenic B cells, we confirmed the downregulation of Bcl2 mRNA and BCL2 protein in pre-malignant B cells after LFPRLR knockdown (Fig. 4f, g).
In contrast to SLE-prone mice, bone marrow B-cell counts in DLBCL-prone mice were significantly increased after LFPRLR knockdown. However, their expression of BCL2 was again significantly reduced (Fig. 4h, i). Together with reduced splenic B-cell numbers (Fig. 4d), this suggests that LFPRLR knockdown impairs the dissemination of the pre-malignant bone marrow B cells into the periphery.
Overall, the PRL-LFPRLR axis induces B-cell lymphocytosis and promotes the expression of TCL1 and BCL2 oncoproteins in pre-malignant B cells, thereby raising the chance of overt B-lymphomagenesis (Fig. 4j).
PRL-LFPRLR signaling maintains overt human B-cell malignancies
Circulating and local PRL has been shown to be elevated in malignant lymphomas55,56. However, the consequences of the local and systemic overproduction of PRL in lymphomas are unknown. We determined whether expression of autocrine/paracrine PRL in tumor samples from patients with DLBCL and/or Burkitt’s lymphoma (BL)57,58,59 was indicative of clinical outcome. Higher-than-median expression of PRL transcript (PRLhigh) significantly associated with lower overall survival in patients with DLBCL/BL, both at diagnosis57 and after standard treatments58,59 (Fig. 5a). Because expression of individual PRLR isoforms in these samples was unavailable, we compared overall survival after separating patients into PRLRhigh and PRLRlow groups based on their median mRNA expression of total PRLR. Total PRLR levels did not correlate with survival of DLBCL/BL patients (Supplementary Fig. 9), suggesting either that local production of autocrine PRL was more important than levels of PRLR expression and/or that some tumors still express some level of SFPRLRs.
a Overall survival probabilities of patients with B-cell lymphomas from 3 datasets (GSE4475, GSE10846, and E-TABM-346) divided into PRLhigh and PRLlow based on the median expression of PRL mRNA. GSE4475 includes 123 DLBCL and 36 Burkitt lymphoma patients at diagnosis. GSE10846 includes samples from 414 DLBCL patients collected after treatment with CHOP (cyclophosphamide, hydroxydaunorubicin, vincristine, and prednisone) or rituximab-CHOP, and E-TABM-346 includes samples from 53 DLBCL patients collected after treatment with rituximab-CHOP or CHOP. HR Hazard Ratio. b, c Expression of PRL mRNA in CD3−CD19+ B cells from healthy donors (n = 27) and B-lymphoblasts of patients with high-grade MYC/BCL2-driven B-ALL (n = 18) by whole RNA sequencing (EGAS00001003266, St. Jude, median ± interquartile range, p-value calculated by Mann–Whitney U test) (b); and in CD3−CD19+ B cells isolated from PBMC of healthy donors (n = 3) and in malignant human B-cell lines (Karpas-422, SU-DHL-6, VAL and OCI-LY18, (c)) by qPCR. d Expression of LFPRLR (red dots) and SFPRLR (blue dots) mRNA in CD3−CD19+ B cells, CD3−CD56+ NK cells and CD19−CD3+ T cells sorted from PBMC of healthy donors (n = 3) and malignant B-cell lines (Karpas-422, SU-DHL-6, VAL and OCI-LY18) by qPCR. e–h MTS assay to compare viability of PBMC isolated from healthy donors after treatment with either control SMO or LFPRLR SMO for 48 h (e), and flow cytometry to compare the viability (% FSChighDAPI−) of MACS-sorted CD3−CD19+ B cells (f), CD3−CD56+ NK cells (g), and CD3+ T cells (h) isolated from PBMC of four healthy donors after treatment with 73 nM of either control SMO or LFPRLR SMO for 48 h. i Viable cell number was assessed using MTS assay in malignant human B-cell lines after treatment with control SMO or LFPRLR SMO for 48 h. j Immunoblotting showing reduction in active phosphorylated STAT3 and global STAT3 levels in three out of four malignant B cell lines treated at their IC50 concentration of LFPRLR SMO for 3, 8, 12 and 48 h. β-actin was used as the loading control in immunoblotting. P values for survival curves were measured by log-rank (Mantel–Cox) test. For all other experiments, p-values were calculated by unpaired t test from 3 independent experiments/biological replicates (±SEM), unless otherwise indicated. qPCR of each biological sample was conducted in three technical replicates. UBB was used as the house-keeping gene in qPCR. For all flow cytometry-based measurements of cell death, the IC50 concentration of the Karpas-422 line was used (73 nM). FPKM = fragments per kilobase of transcript per million mapped reads.
Next, we investigated whether B-lymphoblasts in ALL patients with poorly prognostic MYC/BCL2-driven B-ALL60 express more autocrine PRL than their healthy counterparts. Unlike normal B cells, B cells in B-ALL patients produced autocrine PRL (Fig. 5b). While normal B cells expressed both LF/IF and SFs, 50% of the patients with MYC/BCL2-driven B-ALL expressed only the LF/IFPRLR (Supplementary Fig. 10). Consistent with these findings, unlike MYC/BCL2-driven B-ALL (VAL) and DLBCL (Karpas-422, SU-DHL-6, OCI-LY18) human cell lines, normal B cells of healthy donors did not express autocrine PRL and expressed the counteractive SFPRLRs (Fig. 5c, d). We infer that the upregulation of PRL-LF/IFPRLR signaling (high PRL expression and no SFPRLR) likely contributes to the maintenance of aggressive B-cell malignancies.
The preclinical potential of the LFPRLR SMO was confirmed by showing that LFPRLR knockdown is not toxic to healthy human peripheral blood mononuclear cells (PBMCs) (Fig. 5e). Although normal B cells express more LF than SFPRLRs (Fig. 5d), SFPRLRs do not have to be present in a 1:1 ratio to inhibit the activity of LFPRLRs23,25,26. Moreover, the lack of production of autocrine PRL by normal B cells (Fig. 5b, c) likely renders them insensitive to LFPRLR knockdown (Fig. 5f). Unlike B cells, normal T and NK cells did not express the LFPRLR (Fig. 5d) and, therefore, administration of the LFPRLR SMO was not toxic to these cells (Fig. 5g, h, Supplementary Fig. 11).
We measured malignant B-cell survival after LFPRLR SMO treatment by 3-(4,5-dimethylthiazol-2-yl)−5-(3-carboxymethoxyphenyl)−2-(4-sulfophenyl)−2H-tetrazolium (MTS) assay and flow cytometry. Because malignant B cells produce PRL that can drive their PRLR signaling, we did not add exogenous PRL in our experiments. LFPRLR knockdown killed Karpas-422 and SU-DHL-6 DLBCLs with half maximal inhibitory concentration (IC50) at 60–70 nM of LFPRLR SMO; however, IC50s for the VAL B-ALL and OCI-LY18 DLBCL cell lines were 20–40-fold higher (Fig. 5i, Supplementary Fig. 12). Hence, human B-cell malignancies are differentially sensitive to knockdown of the LF/IFPRLR.
Next, we investigated the mechanisms by which LFPRLR knockdown reduces the fitness of malignant B cells. To determine whether suppression of STAT signaling is a potential mechanism of action of the LFPRLR SMO, we compared expression of the ‘STAT-activating’ LFPRLR and the ‘non-STAT-activating’ IF PRLR23 relative to one another and their absolute expressions before and after LFPRLR knockdown. As expected, expression of both LF and IF PRLR decreased after treatment with LFPRLR SMO. However, LFPRLR expression was ~10–80- fold higher than IF expression in each cell line in the absence of treatment, suggesting that LFPRLR SMO may kill malignant lymphocytes by reducing the opportunity for STAT activation by PRL (Supplementary Fig. 13). As predicted, phosphorylation (activation) and production of STAT3 decreased after LFPRLR knockdown in three out of four malignant B cell lines treated with their cell death inducing IC50 concentrations of LFPRLR SMO (Fig.5j, Supplementary Fig. 14). pSTAT5 was undetectable in all cell lines, although levels of global STAT5 were reduced in some lines after treatment with LFPRLR SMO (Supplementary Fig. 15). Our results are consistent with the recent finding that STAT3, and not STAT5, is activated downstream of PRLR in autoimmune B cells18. We conclude that activation of STAT3 downstream from LFPRLR may play an important role in sustaining of B-cell malignancies.
Knockdown of LFPRLR reduces progression of overt human DLBCL in vivo
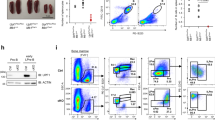
To further demonstrate that knockdown of LFPRLR could be therapeutically beneficial by inhibiting progression of overt B-cell malignancies, we tested the efficacy of LFPRLR SMO in the clearance of two human DLBCL cell-line derived xenografts (CDX) transplanted into immune-deficient NOD-SCID IL2Rγ−/− (NSG) mice. Control SMO or LFPRLR SMO were continuously delivered via alzet minipump for 15 days (Fig. 6a). We confirmed that LFPRLR was significantly knocked down in tumors isolated from mice treated with LFPRLR SMO (Fig. 6b). Consistent with our short-term in vitro studies (Fig. 5i), we observed that LFPRLR knockdown significantly slowed DLBCL progression in vivo in the two independent CDX models used (Fig. 6c, d). We conclude that the knockdown of LFPRLR effectively reduces the progression of overt B-cell malignancies. Because the human LFPRLR SMO does not affect mouse LFPRLR61 we also conclude that this result stems from a direct effect on the B-cell lymphomas. At the same dose of transplanted lymphoma cells, the tumors derived from SU-DHL-6 were more sensitive to LFPRLR knockdown than those derived from OCI-LY18, consistent with the in vitro results (Figs. 5i and 6c, d).
a Experimental design for measuring the in vivo efficacy of LFPRLR SMO in reducing progression of overt human DLBCL in malignant B-cell line derived xenograft (CDX) models. b qPCR validation of LFPRLR knock down in tumors isolated from euthanized transplant recipient mice treated with control SMO (n = 6 for SU-DHL-6 and n = 5 for OCI-LY18, red) or LFPRLR SMO (n = 4 for SU-DHL-6 and n = 6 for OCI-LY18, blue) on day 15. Each dot in qPCR analyses of transcripts corresponds to the mean expression of that gene in tumors from one CDX recipient mouse calculated from 3 technical replicates. UBB was used as the house-keeping gene in qPCR. c, d Change in tumor volume and tumor weight on the day of euthanasia (day 15) in NSG mice that received subcutaneous injections of 7 × 106 malignant human B cells [SU-DHL-6 (c) and OCI-LY18 (d)] and were treated with control SMO (n = 6 for SU-DHL-6 and n = 5 for OCI-LY18, red) or LFPRLR SMO (n = 6 for SU-DHL-6 and n = 6 for OCI-LY18, blue). Tumor volume panels in (c, d) show mean ± SEM. All other graphs show median ± interquartile range. Exact p-values were calculated using the Mann–Whitney U test. ns non-significant, *0.01 < p < 0.05, **0.001 < p < 0.01. a was created with BioRender.com.
Knockdown of LFPRLR in malignant human B cells reduces BCL2 and MYC expression
Given the differential sensitivity of the cell lines to LF/IFPRLR knockdown both in vitro and in vivo, we hypothesized that the intrinsic properties of malignant B cells, including their level of autocrine PRL secretion and expression of oncogenic drivers could dictate their sensitivity to increasing concentrations of LFPRLR SMO.
First, we determined whether differential secretion of PRL from malignant B cells was related to their sensitivity to LFPRLR knockdown. Cells sensitive to nM concentrations of LFPRLR SMO (SU-DHL-6 and Karpas-422, Fig. 5i) secreted much higher levels of PRL than their counterparts sensitive only to μM concentrations (VAL and OCI-LY-18, Fig. 5i) (Fig.7a). Importantly, neutralization of the secreted PRL with rabbit anti-PRL, even at low concentrations, markedly reduced the viability of cell lines with high amounts of PRL secretion and nM sensitivity to LFPRLR SMO (Fig. 7b, Supplementary Fig. 16). In contrast, in malignant B cells sensitive to μM concentrations of LFPRLR SMO, significant impairment of cellular fitness was only observed at high concentrations of anti-PRL (Fig. 7b, Supplementary Fig. 16). Hence, malignant B cells’ dependence on LFPRLR positively correlates with their secretion of autocrine PRL.
a PRL secretion level by ELISA and b viable cell number after treatment with rabbit anti-PRL or rabbit normal serum, assessed by MTS assay in malignant human B-cell lines. P-values by unpaired t test at different concentrations of anti-PRL or normal serum in b: SU-DHL-6 (P = 0.0009 in 1:1600 and P < 0.0001 in 1:800, 1:400, 1:200, 1:100 and 1:50), Karpas-422 (P = 0.0283 in 1:1600, P = 0.0016 in 1:800, P = 0.0004 in 1:200 and P < 0.0001 in 1:400, 1:100 and 1:50), VAL (P = 0.0066 in 1:1600, P = 0.0178 in 1:100 and P = 0.0002 in 1:50) and OCI-LY18 (P = 0.0253 in 1:1600, P = 0.0007 in 1:800, P = 0.0051 in 1:100, P = 0.0007 in 1:50). P-values were calculated by unpaired t test (±SD). P values for 1:50 are indicated in the figure. ns non-significant. c, d LFPRLR mRNA levels and expression of MYC, BCL2 and PRL transcripts in malignant human B-cell lines (Karpas-422, SU-DHL-6, VAL and OCI-LY18) treated with 73 nM of control SMO or LFPRLR SMO for 48 h. Each qPCR sample was run in 3 technical replicates. One representative of 3 independent experiments is shown. UBB was used as the house keeping gene in qPCR. P-values are calculated by unpaired t test from 3 technical replicates (±SEM). Cell lines highly sensitive to low concentration (73 nM) of LFPRLR SMO are shown in (c) and cell lines less sensitive to LFPRLR SMO at 73 nM are shown in d. e, f Immunoblotting to compare the expression of MYC and BCL2 proteins in human DLBCL cell lines (Karpas-422, SU-DHL-6, VAL, and OCI-LY18) treated with 73 nM of control SMO or LFPRLR SMO for 48 h. One representative of 3 independent experiments is shown. β-actin was used as the loading control in immunoblotting. Cell lines highly sensitive to low concentration (73 nM) of LFPRLR SMO are shown in e and cell lines less sensitive to LFPRLR SMO at 73 nM are shown in f. g Signaling of PRL through the LF/IFPRLR induces the expression of MYC and BCL2 in overt human DLBCL and B-ALL, thereby promoting malignant B-cell growth and survival. LFPRLR SMO thus represents a promising candidate to treat these malignancies. g was created with BioRender.com.
We investigated other possible mechanisms that could explain differential sensitivity of malignant B cells to LFPRLR SMO treatment. Of note, treatment with low nanomolar concentrations of LFPRLR SMO significantly reduce LFPRLR expression in SU-DHL-6 and Karpas-422, but not in VAL and OCI-LY18 cell lines. (Fig. 7c, d). These findings suggest dysregulation of the splicing factors required for optimal effect of the LFPRLR SMO in the less sensitive VAL and OCI-LY18 lines.
Splicing dysregulation is often caused by constitutive overexpression of MYC62, an important downstream target of PRL signaling that co-operates with BCL263, another downstream target of PRL16,17,18,19, to sustain B-cell malignancies. A striking difference between the LFPRLR SMO-sensitive SU-DHL-6 and Karpas-422 and the less sensitive VAL and OCI-LY18 lines is the constitutive expression of MYC in addition to BCL2 due to chromosomal translocations in VAL and OCI-LY18. Therefore, we determined whether an inability to downregulate MYC and/or BCL2 in VAL and OCI-LY18 reduced their sensitivity to low concentrations of LFPRLR SMO. As predicted, treatment with a low concentration (73 nM) of LFPRLR SMO reduced MYC protein and BCL2 mRNA and BCL2 protein in the sensitive SU-DHL-6 and Karpas-422 lines, but such reduction was absent in the VAL and OCI-LY18 lines (Fig. 7c–f, Supplementary Fig. 17). We conclude that B-cell malignancies rely on PRL signaling to different extents, with some indication that dual translocations of MYC and BCL2 override this reliance (Fig. 7g).
Discussion
Our newly identified causal role of the PRL-LF/IFPRLR axis in promoting the evolution of B-cell malignancies suggests that the LF/IFPRLR may represent a therapeutic target in patients with B-cell malignancies and in those vulnerable to developing these malignancies.
The molecular mechanisms underlying the increased risk of transformation of B-cell clones in autoimmune diseases are not well understood5,64. We find that LFPRLR not only promotes the accumulation of deleterious B cells by increasing their proliferation and their chances of being mutated by AID, but also exacerbates B-cell pathology in SLE. Thus, targeting the synthesis of the LF/IFPRLR represents an attractive strategy to lower the risk of SLE-associated B-lymphomagenesis.
In pre-malignant B-cells, the LFPRLR promotes expression of BCL2 and TCL1, two oncoproteins whose high levels have been found to correlate with poor clinical prognosis in patients with B-cell lymphomas65,66,67. Hence, our studies underscore the importance of LFPRLR in driving overt B-cell lymphomagenesis. Given the limited attempts to interfere with cancer progression from a pre-malignant state to overt cancer, our studies support the importance of exploring the potential of the LFPRLR SMO in preventing overt leukemogenesis/ lymphomagenesis from pre-leukemic8 and indolent B cells7 and monoclonal B-cell lymphocytosis68.
We find that PRL-LF/IFPRLR signaling maintains the growth and survival of established overt human B-cell malignancies in vitro and in vivo, and that this maintenance is associated with the induction of p-STAT3, MYC, and BCL2. The LFPRLR SMO may thus be a non-toxic alternative to prevent the activation of STAT3, and/or block the production of BCL2 and the ‘difficult-to-drug’ MYC oncoprotein69. Our findings showing that normal T and NK cells are insensitive to LF/IFPRLR knockdown also suggest that LFPRLR SMO may be combined with immune cell-based therapies for treating B-cell malignancies.
The LFPRLR SMO offers several advantages over other drugs that affect PRL signaling. For example, dopamine agonists that inhibit PRL production by the pituitary33,70 have no effect on non-pituitary autocrine/paracrine sources of PRL in the body12, including malignant B cells. Dopamine agonists would also abrogate beneficial PRL-SFPRLR signaling, which may mediate apoptosis of deleterious B cells. By isoform-specific targeting, the chances of off-target toxicities are reduced, such as in the liver, which primarily expresses the SFPRLRs71. Traditional receptor antagonists or anti-PRLR antibodies affect all PRLR isoforms since they all have identical extracellular domains18. Therefore, unlike the LFPRLR SMO, they cannot specifically target the axis governing disease evolution.
Our goal was to determine whether the LFPRLR is causal in B-cell malignancy evolution by examining mouse models exhibiting increased risk of initiation or enhanced progression of this disease, and human data. Each murine model has its limitations. For example, death from SLE in control SMO-treated Mrl-lpr mice27 precludes survival studies that determine the extent to which LFPRLR knockdown blocks the initiation of B-cell lymphomas. In TCL1-tg mice, development of overt DLBCL is slow and uneven28, demanding survival studies with a large number of mice and of more than a year’s duration. Therefore, in both models, we instead examined how LFPRLR promotes the expression of well-known molecular drivers of B-lymphomagenesis specific to the age of the mice at the time of treatment completion with the SMOs. Our findings demonstrating a role for LFPRLR signaling in three types of dysregulated B cells (non-malignant but aberrant, pre-malignant, and malignant) strongly support further preclinical development of the LFPRLR SMO.
Methods
SMO
SMO and control oligomers linked to octaguanidine dendrimers for cell/tissue penetration in vivo, were custom synthesized (Gene Tools, Philomath, Oregon) (Supplementary Table 1a). SMO sequences were designed to bind to the intron 9-exon 10 junction in mouse or human LF/IFPRLR pre-mRNA29.
Animal models
Animal studies were conducted in compliance with Institutional Animal Care and Use Committees (IACUC) at City of Hope and University of California, Riverside. 6-week-old mice female SLE-prone MRL-lpr mice homozygous for the Fas cell surface death receptor mutation (Faslpr) were anesthetized with isoflurane, and Alzet minipumps (Durect, Cupertino, CA) were implanted subcutaneously between the scapulae. Mice were randomly assigned to control SMO or LFPRLR SMO groups and coded by ear punch. Animals in each group were housed individually after pump implantation until wound clips were removed. Alzet pumps that delivered 100 pmoles/h/mouse of either control SMO or LFPRLR SMO were changed after 4 weeks. At week 8 of treatment, two hours before euthanasia, each animal received an intraperitoneal injection of 2.8 mg of the nucleoside analog, 5-ethynyl-2′-deoxyuridine (EdU). 8-week-old male and female DLBCL-prone TCL1-tg mice were implanted with Alzet minipumps and treated with control or LFPRLR SMO for 8 weeks, as described for MRL-lpr mice. Immune-deficient NSG mice were used as transplant recipients in the human B-cell malignancy CDX models. Treatment with SMOs in the NSG CDX-recipient mice was conducted as described above for the primary mouse models of SLE and DLBCL, except that the treatment duration was shorter to ensure tumor size remained within guidelines in the control SMO treated animals. Transplant recipient CDX mice were euthanized following the stringent guidelines on tumor volume and health of mice laid down by IACUC of City of Hope.
Tissue processing
Spleens were placed in Roswell Park Memorial Institute (RPMI) medium containing 40 units/ml DNAse (RPMI+), transferred to a 70 μM cell strainer over a new dish, and mashed with a syringe plunger. Bone marrow was flushed from femurs and tibia with RPMI+ using a syringe with a 25½ G needle into a 70 μM cell strainer. A syringe plunger broke up cell clumps and residual cells were washed through the strainer. After pelleting and red blood cell lysis in 1X lysis buffer (BD Biosciences), WBCs were washed in RPMI+. To ensure that all samples across different groups were treated identically, cells were resuspended in freezing solution (90% FBS/10% DMSO) and stored in liquid nitrogen until analysis.
Cell lines and culture
Mycoplasma-negative human cell lines were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen and cultured in RPMI 1640 with 10% fetal bovine serum, 100 U/mL Penicillin, and 100 μg/mL Streptomycin (Complete RPMI) (Invitrogen/Life technologies). Because human PRLRs do not respond to non-human PRL61, bovine PRL in the FBS-supplemented media would not influence the human lymphoma cell lines used.
Quantitative real-time PCR (qPCR)
RNA was extracted using a kit (NucleoSpin® RNA Plus, MACHERY-NAGEL) according to manufacturer’s instructions. cDNA was synthesized using SuperScript™ IV Reverse Transcriptase (Invitrogen). qPCR was conducted using Power SYBR Green PCR master mix (ThermoFisher Scientific) and QuantStudio 7 flex real-time PCR system (Applied Biosystems). Supplementary Table 1b lists qPCR primers.
Total murine Prlr = LF + SF3 (SF1 and SF2 were not expressed in splenocytes and B cells). Total human PRLR = LF + IF + SF1a + SF1b. There is a huge degree of variability in the absolute expression of the isoforms across individual mice in each strain. To account for this variability and to most accurately predict changes in pro-proliferative, anti-apoptotic effects of PRLRs in cells after LFPRLR knockdown, expression of PRLR isoforms in each mouse was represented as the ratio of LFPRLR: total PRLR. An exception to calculating the ratio is made in instances where the cells express only LF/IFPRLR, as in some malignant human B cells.
B-cell clonality
IGH repertoire of splenic B cells were PCR-amplified using primers for the J558 VH-region gene and the Cμ constant region (Supplementary Table 1c), as described previously72,73. Selective amplification of the J558 variable region of the IGH (VH) family was conducted due to the increased representation of this family in both the production of autoantibodies and non-binding antibodies in MRL-lpr SLE-prone mice74. Amplicons were subjected to next generation sequencing (NGS): TrueSeq library was prepared by adapter ligation to full-length products and sequenced on the MiSeq Illumina 2 × 150 bp platform. Abundances of unique nucleotide sequences were calculated and CDR3 length, spectrum, and functionality were analyzed using International Immunogenetics Information System (IMGT) HighV-QUEST. VDJTools (v1.1.7) was used for visualization. CDR3 amino acid frequencies were calculated using customized R scripts and heatmaps were generated using the pheatmap package and unsupervised hierarchical clustering using Euclidian distance. NGS data are available in GSE207186.
Analysis of PRLR isoforms in B-ALL patients
Expression of human PRLR isoforms was parsed based on previous knowledge that SFs lack most of exon 10, whereas LF and IF include entire or large portions of exon 1021. Raw paired-end fastq files of RNA-seq were mapped to human GRCh38 using STAR v.2.7.6a. Raw exon counts across the whole genome were obtained using DEXSeq75, normalized based on variance stabilizing transformation method using R package DESeq276, and batch corrected using R package sva.
Flow cytometry
Cells were thawed in Complete RPMI and stained with fluorochrome-tagged surface antibodies and Ghost-UV450 for 30 min on ice. Using eBioscience™ Transcription Factor Staining Buffer Set, cells were fixed, permeabilized, and stained with intracellular antibodies for 30 min on ice followed by acquisition on the BD FACSymphony cytometer. Gates were set using single-stained and fluorescence minus one (FMO) controls. Data were analyzed using FlowJo10.7.1. Supplementary Table 2a lists cytometry antibodies.
Cell cycle
EdU incorporation was assessed using Click-iT™ Plus EdU flow cytometry. Cells were washed in 1% bovine serum albumin in phosphate buffered saline and stained for surface antigens, as above, followed by fixation, permeabilization, click-iT EdU detection, and 4′,6-diamidino-2-phenylindole (DAPI) staining. Cells were analyzed on BD FACSymphony and FlowJo10.7.1.
Magnetic activated cell sorting (MACS)
Mouse splenic B cells were enriched using CD19-microbeads (Miltenyi). Human healthy donor peripheral blood mononuclear B, T, and NK cells (PBMCs) were enriched using Miltenyi kits. Enrichment was confirmed by flow cytometry.
Immunoblotting
Cells were lysed in RIPA buffer (Sigma) supplemented with 1% protease inhibitor ‘cocktail’ (Pierce). Proteins were separated by electrophoresis through 4–20% TGX gradient gels (BioRad) and transferred to polyvinylidene fluoride membranes (Immobilon; Millipore). Mouse and human proteins were detected by immunoblotting using antibodies (Supplementary Table 2b) and western enhanced chemiluminescence (BioRad).
MTS Assay
One thousand cells were seeded per well in 96-well plates and incubated in Complete RPMI containing different concentrations of control or LFPRLR SMO (0, 5, 25, 125, 250, 500, 750, 1000, 1500, 2000 nM) at 37 °C for 48 h. Twenty μl of MTS (Promega) was added to 100 μl of media containing cells and incubated for 3 h. Absorbance was measured at 490 nm with TECAN Infinite M1000 Pro microplate reader.
Enzyme linked immunosorbent assays (ELISA)
For the quantitative determination of human PRL concentrations in cell supernatant, we used Biotechne Quantikine ELISA kit Cat #DPRL00. According to manufacturer’s instructions, we measured secreted PRL in cells supernatant based on standard curve plotted using different concentrations of human recombinant PRL. Sensitivity of human PRL ELISA kit is 0.264 ng/mL.
Anti-dsDNA was assayed by ELISA using a kit from Chondrex (#3031). To ensure consistent values between assays, we diluted and then stored samples in assay dilution buffer for >24 h at −20 °C before assay.
PRL neutralization assay
Ten thousand cells were seeded per well in 96-well plates and incubated in Complete RPMI containing different concentrations of normal rabbit serum or rabbit anti-PRL (NIDDK standard, AFP55762089) (0, 1:1600, 1:800, 1:400, 1:200, 1:100, 1:50 dilution) at 37 °C for 48 h. Twenty μl of MTS (Promega) was added to 100 μl of media containing cells and incubated for 3 h. Absorbance was measured at 490 nm with a TECAN Infinite M1000 Pro microplate reader.
LFPRLR SMO treatment efficacy studies in CDX models of human B-cell malignancies
Six female immune-deficient NSG mice (Jackson Laboratories) received subcutaneous injections of 7 × 106 malignant B cells suspended in 200 μL matrigel (BD Biosciences) in their right flank. Cell lines were pre-treated in vitro for 3 h with either control SMO or LFPRLR SMO at their respective IC50 concentrations. Alzet minipumps delivering the SMOs were implanted on the left scapula of each mouse. Tumor burden was assessed twice weekly by caliper measurement. Tumor volume was calculated using the formula: V = ½ (length × width2). Mice were euthanized after 15 days of treatment.
Statistics and reproducibility
Exact p-values are provided if significant (P < 0.05) or trending towards significance (0.05 < P < 0.1). Statistical tests were two-tailed. Pairwise comparisons between mouse cohorts were performed using the Mann–Whitney U test. For Kaplan–Meier survival analyses, p-values were calculated using a log-rank test. Female SLE-prone mice were used because SLE is more frequent in females. In experiments involving pre-malignant or malignant B cells, we represented both sexes equally because B-cell malignancies develop at the same frequencies in both males and females and malignant B cells from males and females had similar PRL and PRLR isoform expression. qPCR samples were run in 3 technical replicates. Experiments involving cell lines were repeated 3 times to determine reproducibility.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Next generation sequencing of the immunoglobulin heavy chain repertoire described in this study is available in Gene Expression Omnibus under accession GSE207186. All source data for main figures in the manuscript are provided as supplementary Data 1. Uncropped and unedited immunoblots are provided in Supplementary Figures.
References
Sinha, R., Nastoupil, L. & Flowers, C. R. Treatment strategies for patients with diffuse large B-cell lymphoma: past, present, and future. Blood Lymphat Cancer 2012, 87 (2012).
Paul, S., Kantarjian, H. & Jabbour, E. J. Adult acute lymphoblastic leukemia. Mayo Clin. Proc. 91, 1645–1666 (2016).
Bernatsky, S. et al. Cancer risk in childhood-onset systemic lupus. Arthritis Res. Ther. 15, 1–4 (2013).
Bernatsky, S. et al. Lymphoma risk in systemic lupus: effects of disease activity versus treatment. Ann. Rheum. Dis. 73, 138–142 (2014).
Koff, J. L. & Flowers, C. R. B cells gone rogue: the intersection of diffuse large B cell lymphoma and autoimmune disease. Expert Rev. Hematol. 9, 553–561 (2016).
White, C. A. et al. AID dysregulation in lupus-prone MRL/Fas lpr/lpr mice increases class switch DNA recombination and promotes interchromosomal c-Myc/IgH loci translocations: modulation by HoxC4. Autoimmunity 44, 585–598 (2011).
Lossos, I. S. & Gascoyne, R. D. Transformation of follicular lymphoma. Best. Pract. Res. Clin. Haematol. 24, 147–163 (2011).
Mori, H. et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc. Natl Acad. Sci. 99, 8242–8247 (2002).
Swaminathan, S. et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat. Immunol. 16, 766–774 (2015).
Bernard, V., Young, J. & Binart, N. Prolactin—a pleiotropic factor in health and disease. Nat. Rev. Endocrinol. 15, 356–365 (2019).
Abramicheva, P. & Smirnova, O. Prolactin receptor isoforms as the basis of tissue-specific action of prolactin in the norm and pathology. Biochem. (Mosc.) 84, 329–345 (2019).
Ben-Jonathan, N., Liby, K., McFarland, M. & Zinger, M. Prolactin as an autocrine/paracrine growth factor in human cancer. Trends Endocrinol. Metab. 13, 245–250 (2002).
Peeva, E. & Zouali, M. Spotlight on the role of hormonal factors in the emergence of autoreactive B-lymphocytes. Immunol. Lett. 101, 123–143 (2005).
Saha, S., Tieng, A., Pepeljugoski, K. P., Zandamn-Goddard, G. & Peeva, E. Prolactin, systemic lupus erythematosus, and autoreactive B cells: lessons learnt from murine models. Clin. Rev. Allergy Immunol. 40, 8–15 (2011).
Walker, S. E., Allen, S. H., Hoffman, R. W. & McMurray, R. W. Prolactin: a stimulator of disease activity in systemic lupus erythematosus. Lupus 4, 3–9 (1995).
Berczi, I., Nagy, E, De Toledo, S., Matusik, R. & Friesen, H. Pituitary hormones regulate c-myc and DNA synthesis in lymphoid tissue. J. Immunol. 146, 2201–2206 (1991).
Dominguez-Caceres, M. A. et al. Prolactin induces c-Myc expression and cell survival through activation of Src/Akt pathway in lymphoid cells. Oncogene 23, 7378–7390 (2004).
Flores-Fernández, R. et al. Prolactin rescues immature B cells from apoptosis-induced BCR-aggregation through STAT3, Bcl2a1a, Bcl2l2, and Birc5 in lupus-prone MRL/lpr mice. Cells 10, 316 (2021).
Kochendoerfer, S., Krishnan, N., Buckley, D. & Buckley, A. Prolactin regulation of Bcl-2 family members: increased expression of bcl-xL but not mcl-1 or bad in Nb2-T cells. J. Endocrinol. 178, 265–274 (2003).
Walker, A. M. et al. Prolactin-immunoglobulin G complexes from human serum act as costimulatory ligands causing proliferation of malignant B lymphocytes. Proc. Natl Acad. Sci. 92, 3278–3282 (1995).
Trott, J., Hovey, R., Koduri, S. & Vonderhaar, B. Alternative splicing to exon 11 of human prolactin receptor gene results in multiple isoforms including a secreted prolactin-binding protein. J. Mol. Endocrinol. 30, 31–48 (2003).
Ormandy, C. J., Binart, N., Helloco, C. & Kelly, P. A. Mouse prolactin receptor gene: genomic organization reveals alternative promoter usage and generation of isoforms via alternative 3′-exon splicing. DNA Cell Biol. 17, 761–770 (1998).
Grible, J. M. et al. The human intermediate prolactin receptor is a mammary proto-oncogene. NPJ Breast Cancer 7, 1–11 (2021).
Kavarthapu, R. & Dufau, M. L. Prolactin receptor gene transcriptional control, regulatory modalities relevant to breast cancer resistance and invasiveness. Front. Endocrinol. 13, 949396 (2022).
Tan, D. & Walker, A. M. Short form 1b human prolactin receptor down-regulates expression of the long form. J. Mol. Endocrinol. 44, 187 (2010).
Van Coppenolle, F. et al. Prolactin stimulates cell proliferation through a long form of prolactin receptor and K+ channel activation. Biochem. J. 377, 569–578 (2004).
Andrews, B. S. et al. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J. Exp. Med. 148, 1198–1215 (1978).
Hoyer, K. K. et al. Dysregulated TCL1 promotes multiple classes of mature B cell lymphoma. Proc. Natl Acad. Sci. 99, 14392–14397 (2002).
Yonezawa, T. et al. Anti-metastatic outcome of isoform-specific prolactin receptor targeting in breast cancer. Cancer Lett. 366, 84–92 (2015).
Chen, K.-H. E. et al. Prolactin enhances T regulatory cell promotion of breast cancer through the long form prolactin receptor. Transl. Oncol. 14, 101195 (2021).
Rys, R. N., Venkataraman, M., Zeng, J., Mann, K. K. & Johnson, N. J. B. Fas Mutations in Non-Hodgkin’s Lymphoma (NHL): implications for disease progression and therapeutic resistance. Blood 134, 1520 (2019).
Grønbæk, K. et al. Somatic Fas mutations in non-Hodgkin’s lymphoma: association with extranodal disease and autoimmunity. Blood 92, 3018–3024 (1998).
Tsokos, G. C. Systemic lupus erythematosus. N. Engl. J. Med. 365, 2110–2121 (2011).
Malkiel, S., Barlev, A. N., Atisha-Fregoso, Y., Suurmond, J. & Diamond, B. Plasma cell differentiation pathways in systemic lupus erythematosus. Front. Immunol. 9, 427 (2018).
Vincent, F. B., Morand, E. F., Schneider, P. & Mackay, F. The BAFF/APRIL system in SLE pathogenesis. Nat. Rev. Rheumatol. 10, 365–373 (2014).
Bekeredjian-Ding, I. B. et al. Plasmacytoid dendritic cells control TLR7 sensitivity of naive B cells via type I IFN. J. Immunol. 174, 4043–4050 (2005).
Cerutti, A., Qiao, X. & He, B. Plasmacytoid dendritic cells and the regulation of immunoglobulin heavy chain class switching. Immunol. Cell Biol. 83, 554–562 (2005). J. I. & biology, c.
Ding, C. et al. Plasmacytoid dendritic cells regulate autoreactive B cell activation via soluble factors and in a cell-to-cell contact manner. J. Immunol. 183, 7140–7149 (2009).
Huang, X., Dorta-Estremera, S., Yao, Y., Shen, N. & Cao, W. Predominant role of plasmacytoid dendritic cells in stimulating systemic autoimmunity. Front. Immunol. 6, 526 (2015).
William, J., Euler, C., Christensen, S. & Shlomchik, M. J. J. S. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science 297, 2066–2070 (2002).
William, J., Euler, C. & Shlomchik, M. J. Short-lived plasmablasts dominate the early spontaneous rheumatoid factor response: differentiation pathways, hypermutating cell types, and affinity maturation outside the germinal center. J. Immunol. 174, 6879–6887 (2005).
Wang, X. & Xia, Y. Anti-double stranded DNA antibodies: origin, pathogenicity, and targeted therapies. Front. Immunol. 10, 1667 (2019).
Carreón-Talavera, R. et al. Prolactin promotes proliferation of germinal center B cells, formation of plasma cells, and elevated levels of IgG3 anti-dsDNA autoantibodies. Front. Immunol. 13, https://doi.org/10.3389/fimmu.2022.1017115 (2022).
McDonnell, T. J. et al. Deregulated Bcl-2-immunoglobulin transgene expands a resting but responsive immunoglobulin M and D-expressing B-cell population. Mol. Cell Biol. 10, 1901–1907 (1990).
Kipps, T. J. & Carson, D. A. Autoantibodies in chronic lymphocytic leukemia and related systemic autoimmune diseases. Blood 81, 2475–2475 (1993).
Wardemann, H. et al. Predominant autoantibody production by early human B cell precursors. Science 301, 1374–1377 (2003).
Johnson, T. A., Rassenti, L. Z. & Kipps, T. J. Ig VH1 genes expressed in B cell chronic lymphocytic leukemia exhibit distinctive molecular features. J. Immunol. 158, 235–246 (1997).
Saini, S. S., Allore, B., Jacobs, R. M. & Kaushik, A. Exceptionally long CDR3H region with multiple cysteine residues in functional bovine IgM antibodies. Eur. J. Immunol. 29, 2420–2426 (1999).
Tsai, A. G. et al. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell 135, 1130–1142 (2008).
Muramatsu, M. et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 (2000).
Mai, T. et al. Estrogen receptors bind to and activate the HOXC4/HoxC4 promoter to potentiate HoxC4-mediated activation-induced cytosine deaminase induction, immunoglobulin class switch DNA recombination, and somatic hypermutation. J. Biol. Chem. 285, 37797–37810 (2010).
Merino, R. N., Ding, L., Veis, D. J., Korsmeyer, S. J. & Nuñez, G. Developmental regulation of the Bcl‐2 protein and susceptibility to cell death in B lymphocytes. EMBO J. 13, 683–691 (1994).
Keenan, R. A. et al. Censoring of autoreactive B cell development by the pre-B cell receptor. Science 321, 696–699 (2008).
Osmond, D. G. Proliferation kinetics and the lifespan of B cells in central and peripheral lymphoid organs. Curr. Opin. Immunol. 3, 179–185 (1991).
Grobe, N., Rühle, H. & Eckelmann, G. Hyperprolactinämie bei malignen Lymphomen. DMW-Dtsch. Med. Wochenschr. 115, 1825–1827 (1990).
Matera, L. et al. Expression of prolactin and prolactin receptors by non‐Hodgkin’s lymphoma cells. Int. J. Cancer 85, 124–130 (2000).
Hummel, M. et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 354, 2419–2430 (2006).
Lenz, G. et al. Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 359, 2313–2323 (2008).
Jais, J.-P. et al. The expression of 16 genes related to the cell of origin and immune response predicts survival in elderly patients with diffuse large B-cell lymphoma treated with CHOP and rituximab. Leukemia 22, 1917–1924 (2008).
Gu, Z. et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 51, 296–307 (2019).
Utama, F. E. et al. Insensitivity of human prolactin receptors to nonhuman prolactins: relevance for experimental modeling of prolactin receptor-expressing human cells. Endocrinology 150, 1782–1790 (2009).
De Kesel, J., Fijalkowski, I., Taylor, J. & Ntziachristos, P. Splicing dysregulation in human hematologic malignancies: beyond splicing mutations. Trends Immunol. 43, 674–686 (2022).
Fairlie, W. D. & Lee, E. F. Co-operativity between MYC and BCL-2 pro-survival proteins in cancer. Int. J. Mol. Sci. 22, 2841 (2021).
Tessier-Cloutier, B. et al. Cell of origin in diffuse large B-cell lymphoma in systemic lupus erythematosus: molecular and clinical factors associated with survival. Lupus Sci. Med. 6, e000324 (2019).
Hu, S. et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. J. Am. Soc. Hematol. 121, 4021–4031 (2013).
Aggarwal, M. et al. TCL1A expression delineates biological and clinical variability in B-cell lymphoma. Mod. Pathol. 22, 206–215 (2009).
Ramuz, O. et al. Identification of TCL1A as an immunohistochemical marker of adverse outcome in diffuse large B-cell lymphomas. Int. J. Oncol. 26, 151–157 (2005).
Strati, P. & Shanafelt, T. D. Monoclonal B-cell lymphocytosis and early-stage chronic lymphocytic leukemia: diagnosis, natural history, and risk stratification. Blood J. Am. Soc. Hematol. 126, 454–462 (2015).
Dang, C. V., Reddy, E. P., Shokat, K. M. & Soucek, L. Drugging the’undruggable’cancer targets. Nat. Rev. Cancer 17, 502–508 (2017).
Walker, S. Treatment of systemic lupus erythematosus with bromocriptine. Lupus 10, 197–202 (2001).
Nagano, M. & Kelly, P. A. Tissue distribution and regulation of rat prolactin receptor gene expression. Quantitative analysis by polymerase chain reaction. J. Biol. Chem. 269, 13337–13345 (1994).
Swaminathan, S. et al. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat. Med. 19, 1014–1022 (2013).
Duy, C. et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J. Exp. Med. 207, 1209–1221 (2010).
Foster, M. H., MacDonald, M., Barrett, K. J. & Madaio, M. P. VH gene analysis of spontaneously activated B cells in adult MRL-lpr/lpr mice. J558 bias is not limited to classic lupus autoantibodies. J. Immunol. 147, 1504–1511 (1991).
Anders, S., Reyes, A. & Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Acknowledgements
We thank Dr. Alexey Danilov (City of Hope) for providing us with human DLBCL and B-ALL cell lines. We thank Dr. Nora Heisterkamp and Dr. Rui Su (City of Hope) for their productive discussions. Schematics were created using BioRender.com.
This work was supported by the following grants: City of Hope-University of California Riverside Biomedical Research Initiative Award (S.S., A.M.W.), Research Start-Up Budget from the Beckman Research Institute of the City of Hope (S.S.), Junior Investigator Research Development Award from the Circle of Service Foundation (S.S.), P50 CA107399-12 City of Hope Lymphoma SPORE Career Enhancement Program Pilot Award (S.S.) from the National Cancer Institute of the National Institutes of Health [PIs: Stephen J. Forman (S.J.F.), Larry Kwak], a Shared Resources Pilot Grant from City of Hope (S.S.), a Conquer Cancer Now Award from the Concern Foundation (S.S.), and a Congressional Families Program Award from the Prevent Cancer Foundation (S.S.). S.S. is a Scholar of the American Society of Hematology and is supported in part by a Translational Research Program Grant (LLS 6624-21) from the Leukemia and Lymphoma Society. Research reported in this publication included work performed in the Analytical Cytometry Core and Integrative Genomics Core Shared Resource supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
A.T.K. conducted most of the experiments, analyzed data, and interpreted the results. A.K. assisted A.T.K. with flow cytometry experiments, data analyses, and interpretation. A.S.O., K.C.R., S.A., S.J.L., A.M.W., and M.Y.L. conducted experiments. Z.H., B.D., and Z.G. developed scripts to parse the expression of specific PRLR isoforms in RNA sequencing data from patients with high-risk B-ALL. X.W. developed scripts to analyze next generation sequencing data of the B-cell repertoire in SLE-prone mice. W.S., I.S., and E.M. provided guidance on B-cell pathology in SLE. M.M., S.J.F., and J.L.K. provided scientific advice in B-cell malignancy pathogenesis and transition of autoimmune diseases to B-cell malignancies. A.M.W. provided expert advice on PRL signaling. S.S. and A.M.W. conceived the study, developed the experimental methodology, interpreted the results, provided administrative, technical, and material support and supervised the study. S.S. wrote the manuscript. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.M.W. and S.S. declare the following competing interests: The work described in this study is covered under a pending US patent application (Inventors: A.M.W. and S.S.). All other authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Guideng Li and Eve Rogers. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Taghi Khani, A., Kumar, A., Sanchez Ortiz, A. et al. Isoform-specific knockdown of long and intermediate prolactin receptors interferes with evolution of B-cell neoplasms. Commun Biol 6, 295 (2023). https://doi.org/10.1038/s42003-023-04667-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-023-04667-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.