Abstract
Non-alcoholic fatty liver disease (NAFLD) is associated with hepatic metabolism dysfunction. However, the mechanistic role of miR204 in the development of NAFLD is unknown. We investigate the functional significance of miR204 in the evolution of NAFLD. IDH2 KO mice feed a normal diet (ND) or HFD increased body weight, epididymal fat-pad weight, lipid droplet in liver, blood parameter and inflammation compared to WT mice fed a ND or HFD. Moreover, the expression of miR204 is increased in mice with IDH2 deficiency. Increased miR204 by IDH2 deficiency regulates carnitine palmitoyltransferase 1a (cpt1a) synthesis, which inhibits fatty acid β-oxidation. Inhibition of miR204 prevents the disassembly of two fatty acid-related genes by activating CPT1a expression, which decreases lipid droplet in liver, inflammatory cytokines, epididymal fat pad weight, blood parameters. Increased miR204 by IDH2 deficiency promotes the pathogenesis of HFD-induced NAFLD by regulating hepatic fatty acid metabolism and inflammation.
Similar content being viewed by others
Introduction
Metabolic disorders are rapidly increasing in incidence due to higher rates of obesity, diabetes, hyperlipidemia, and cardiovascular disease1,2. Non-alcoholic fatty liver disease (NAFLD) is a liver condition not caused by heavy alcohol consumption; its main characteristic is excess fat storage in liver cells. NAFLD is emerging as a major cause of chronic liver disease worldwide, accounting for 20% of cases globally3. Some patients with NAFLD develop non-alcoholic steatohepatitis (NASH), which is an aggressive form of fatty liver disease induced by liver inflammation that may progress to liver failure and cancer4.
Imbalance of energy homeostasis causes dysfunction of muscles, liver, and adipocytes, leading to metabolic disease. To remove excess energy, it is necessary to increase metabolic activity via fatty acid β-oxidation. Promoting the activation of lipid metabolism in mitochondria is essential5,6; mitochondrial dysfunction causes obesity-related disorders, but the underlying mechanisms are unclear7,8.
The mitochondrial protein, isocitrate dehydrogenase (IDH2) is essential for energy consumption, and converts the product of oxidative decarboxylation of isocitrate into α-ketoglutarate along with reduction of NADP+ to NADPH. IDH2 deficiency increases mitochondrial dysfunction and endothelial cell inflammation9,10. IDH2 deficiency induces the accumulation of citrate, which is transported from mitochondria to the cytosol11. We hypothesized that cytosol citrate could be converted into Acetyl-CoA, which is a precursor of fatty acids and cholesterol. IDH2 protects against the metabolic stress induced by a high-fat diet by limiting oxidative damage to the mitochondria in brown adipose tissue12. IDH2 knockout (KO) mice exhibit obesity resistance and insulin sensitivity13. However, there are no previous studies related to IDH2 deficiency and fat metabolism in the liver, where nutrients from the hepatic portal vein are synthesized into fat. Here, we examined the role of IDH2 deficiency in lipid metabolic disorder.
Carnitine palmitoyltransferase 1a (CPT1a) is a transporter of fatty acids for β-oxidation. This mitochondrial enzyme mediates the degradation of accumulated fatty acids for energy production in mitochondria. A decreased CPT1a level blocks fatty acid degeneration in mitochondria, resulting in intracellular accumulation of long-chain fatty acids. CPT1a overexpression protected against high-fat diet (HFD)-induced weight gain, insulin resistance, endoplasmic reticulum stress, inflammation, and liver damage14.
MicroRNAs (miRNAs) are small non-coding RNAs that regulate target gene expression and translation via binding to target mRNAs. miRNAs are expressed in various diseases, including metabolic disorders15,16. miR204 is abundant in damaged pancreatic β-cells17; however, whether it regulates metabolic disorders and obesity in vivo is unknown. miR204 regulates many functionally important β-cell genes. miR204 inhibition attenuates the HFD-triggered imbalance in glucose uptake18,19. We analyzed the hepatic miRNA profiles of IDH2 KO mice and wild-type (WT) mice. miR204 expression was more than threefold higher in IDH2 KO mice compared to WT mice.
In this study, we showed that miR204 was upregulated in steatotic hepatocytes and the livers of IDH2 KO mice as a NAFLD model. Elevation of miR204 in vitro and in vivo induced lipid accumulation and fatty acid synthesis in hepatocytes by destabilizing cpt1a mRNA to inhibit fatty acid β-oxidation. A miR204 inhibitor (miR204-I) significantly attenuated body weight gain, subcutaneous fat accumulation, and hepatic inflammation. Our findings reveal a novel post-transcriptional mechanism by which miR204 regulates CPT1a expression in the pathogenesis of NAFLD.
Results
IDH2 deficiency promotes HFD-induced NAFLD with obesity and inflammation in the liver
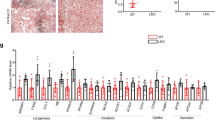
To investigate the role of IDH2 in metabolic diseases characterized by tricarboxylic acid cycle (TCA) disorders, we compared the body weights of IDH2 KO and WT mice fed a normal diet (ND) or HFD. IDH2 KO mice fed an ND or HFD showed significantly increased body weight compared to WT mice (Fig. 1a, b). IDH2 KO mice fed an ND or HFD had epididymal fat pads about twice as large as those of WT mice (Fig. 1c). IDH2 KO mice showed significantly elevated serum levels of total cholesterol, triglycerides, and low-density lipoprotein (LDL) (Fig. 1d–f). Therefore, IDH2 KO mice fed an ND or HFD exhibited enhanced fat accumulation and cholesterol synthesis compared to WT mice.
a Body weights of male mice fed a normal diet (ND, black-WT, green-KO) and high-fat diet (HFD, blue-WT, red-KO) for 1–12 weeks. b Photographs of WT and KO fed an ND or HFD. c Epididymal fat mass. d–f Blood parameters. Data are means ± SEM of three independent experiments. *p < 0.05 vs. WT-ND mice. #p < 0.05 vs. WT-HFD mice. g Representative H&E-stained high-magnification images of mouse livers. h Representative immunohistochemical images of CD68 in the liver of WT and KO mice fed an ND or HFD; scale bars, 50 μm. i Western blot analysis of CD68 in the liver tissues of two mice per group. j–l Expression of IL-1β (j), IL-6 (k), and TNF-α (l) by quantitative real-time PCR, normalized to GAPDH. Data are means ± SEM of three independent experiments (n = 3). *p < 0.05 vs. WT-ND mice. #p < 0.05 vs. WT-HFD mice.
Fatty acids absorbed in the small intestine are transported to the liver, where they are metabolized and used as an energy source, but excess fat intake induces metabolic disorders by causing hepatic fat accumulation. In addition, given the close relationships among obesity, abnormal lipid metabolism, and inflammation, we investigated the effect of IDH2 deficiency on lipid accumulation and inflammation in the liver. Histological analysis was performed to determine whether IDH2 deficiency induces metabolic disease. Hematoxylin-eosin (H&E) staining of liver sections showed greater hepatic lipid droplet accumulation in HFD-fed IDH2 KO mice compared to HFD-fed WT mice, and the same pattern was observed in ND-fed mice (Fig. 1g). Obesity contributes to activation and recruitment of inflammatory cells such as monocytes, neutrophils, and macrophages20. Chronic inflammation is responsible for the side effects of several metabolic diseases. Immunohistochemical staining for the macrophage marker CD68 was performed to assay macrophage infiltration in liver tissue. CD68 expression was significantly increased in HFD-fed IDH2 KO mice compared to HFD-fed WT mice (Fig. 1h). This finding was confirmed by western blotting (Fig. 1i). Under inflammatory conditions, macrophages secrete cytokines such as interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF)-α21, causing vascular permeability and recruitment of inflammatory cells. The cytokine levels were increased in IDH2 KO mice (Fig. 1j–l). Therefore, IDH2 deficiency induces fat accumulation and an inflammatory response in the liver.
IDH2 deficiency upregulates fatty acid and cholesterol synthesis
IDH2 is a mitochondrial enzyme involved in the TCA cycle. In the absence of IDH2, citrate is not converted into α-KG in mitochondria. It was hypothesized that accumulated citrate exits the cytoplasm via channels and is converted into acetyl-CoA, thus promoting lipid synthesis (Fig. 2a). We evaluated the citrate and acetyl-CoA levels in liver tissue from IDH2 KO mice fed the ND or HFD (Fig. 2b, c). We also analyzed the mRNA levels of enzymes involved in lipid synthesis using acetyl-CoA as a precursor in IDH2 KO mice. The mRNA levels of ATP binding cassette subfamily G member 1 (ABCG1), cytochrome P450 family 7 subfamily A member 1 (CYP7A1), 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase (HMCGR), and sterol regulatory element-binding protein 1c (SREBP-1c), which are involved in cholesterol synthesis, and acetyl-CoA carboxylase (ACCα), fatty acid synthase (FAS), and peroxisome proliferator-activated receptor γ (PPARγ), which are involved in fatty acid synthesis, were increased in IDH2 KO mice fed the ND or HFD. The mRNA levels of acyl-coenzyme A oxidase 1 (ACOX-1), long-chain acyl-CoA dehydrogenase (LCAD), peroxisome proliferator-activated receptor alpha (PPARα), pyruvate dehydrogenase kinase 4 (PDK4), and uncoupling protein 2 (UCP2), which are involved in fatty acid β-oxidation, were decreased in IDH2 mice compared to WT mice fed the ND or HFD (Fig. 2d, e). In summary, IDH2 KO results in accumulation of acetyl-CoA, which is converted into fatty acids and cholesterol, thereby suppressing fatty acid β-oxidation.
a Mechanisms of fatty acid and cholesterol synthesis. b, c Citrate and acetyl-CoA levels in liver tissue determined using a colorimetric/fluorometric assay kit. d, e Expression of genes related to cholesterol efflux and synthesis, and fatty acid synthesis and β-oxidation, revealed by quantitative real-time PCR (normalized to GAPDH). Data are means ± SEM of three independent experiments (n = 3). *p < 0.05 vs. WT-ND mice. #p < 0.05 vs. WT-HFD mice.
IDH2 deficiency increases miR204 expression in the liver
miRNAs are implicated in the development of NAFLD22,23. NAFLD is closely related to obesity, diabetes, and insulin resistance. Therefore, we investigated miRNA expression in HFD-induced obesity. The livers of WT and IDH2 KO mice were analyzed using the miRNA 4.0 array (Affymetrix, Santa Clara, CA, USA) by Macrogen (Seoul, South Korea). In total, 38 of 3,163 miRNAs were significantly (at least threefold) differentially regulated between the livers of WT and IDH2 KO mice (Fig. 3a). Among the 38 miRNAs, 8 miRNAs are reportedly associated with metabolic and cardiovascular diseases (miR-802-3p, miR125b-2-3p, miR714, miR700-5p, miR148a-5p, miR204-5p, miR-188-5p, and miR451a). We arranged the fold values of eight miRNAs in IDH2 KO mice and WT mice (Fig. 3b). To validate the sequencing data, the expression of these eight miRNAs was measured by quantitative PCR (qPCR) in the livers of WT and IDH2 KO mice from 10 mice in each group (Fig. 3c). We chose miR204 because of higher accuracy in these results, as compared to the sequencing data in Fig. 3a, b due to more number of mice used in the experiment. Generally, miRNAs regulate target gene expression by binding to the mRNA 3′UTR. The seed site is important for binding a miRNA to an mRNA. The seed site (or seed region) is a conserved heptametric sequence in mammals, which is typically situated 2–7 nucleotides from the 5′-end of the miRNA. miRNA significantly affects target gene mRNA expression in the 3′UTR. Although it is less effective than matching the 3′-UTR sequence, coding sequence (CDS) matching of miRNAs suppresses mRNA translation24. The top three miRNA-target genes (miR188-5p, miR204, and miR451a) were analyzed and matched to the CDS and 3′-UTR of genes with no mRNA alterations (stearoyl-CoA desaturase [SCD1], CPT1a, and medium-chain acyl-coenzyme [MCAD]) (Table 1). miR204 expression was increased in HFD-fed WT mice and IDH2 KO mice (Fig. 3d). Correlation of lipid accumulation and miR204 expression was detected through counting of droplet in H&E staining with miR204 qPCR results. As a result, we confirmed that there was a positive correlation using Pearson’s correlation coefficient between lipid accumulation and miR204 expression, and in the case of a high-fat diet, IDH2 deficiency further enhanced the correlation (Fig. 3e). We confirmed miR204 expression using miR204 inhibitor to confirm the association between miR204 and lipid synthesis also in vitro. An increase in miR204 was confirmed in fatty acid-treated hepatocytes (Fig. 3f), and as a result of controlling the expression of miR204 via a miR204 inhibitor, fat staining with oil red O decreased (Fig. 3g). Taken together, miR204 expression and lipid accumulation have a positive correlation, and that this relationship is further strengthened when IDH2 deficiency occurs.
a In total, 38 of 3163 miRNAs showed significant (at least threefold) up or downregulation. b Arrangement of fold values of eight miRNA array results reportedly linked to metabolic and cardiovascular diseases in IDH2 KO mice and WT mice liver. c Repeated by qPCR in the livers of WT and IDH2 KO mice from ten mice in each group d miR204 expression in mouse liver tissues revealed by qPCR. e Positive correlation of lipid accumulation and miR204 expression in liver. Pearson correlation coefficient (WT_ND: 0.51667, IDH2 KO_ND:0.28901096, WT_HFD:0.67427449, IDH2 KO_HFD:0.73240585). These cells were first transfected siCON, siIDH2 and miR204-I after 24 h, and then treated fatty acid 24 h. f miR204 expression in hepatocytes; RNU6 was used as the internal control. g Oil Red O staining of hepatocytes. Scale bars, 20 μm. Data are means ± SEM of ten independent experiments (n = 10). *p < 0.05 vs. WT-ND mice or control hepatocyte. #p < 0.05 vs. WT-HFD mice or siIDH2-transfected hepatocyte.
miR204 inhibits the translation of CPT1a in vitro and in vivo
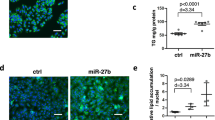
To confirm direct targeting of CPT1a by miR204, we ectopically expressed the full-length CPT1a cDNA fused with a Myc-tag in hepatocytes. The CPT1a WT or mutated plasmid contained the predicted miR204 binding site (Fig. 4a). We used the cpt1a plasmid and a Homo sapiens-derived plasmid. The target sequence of miR204 was conserved and matched the seed site of the target mRNA of Mus musculus. CPT1a WT or the mutated plasmid was co-transfected with a miR204 mimic (204-M). Overexpression of miR204 inhibited Myc-CPT1a expression in the CPT1a WT plasmid only, but this effect was not seen with mutant plasmid transfection (Fig. 4b, c). A miR204 inhibitor rescued CPT1a expression, which was reduced by IDH2 deletion in ND- or HFD-fed mouse liver tissues and hepatocytes (Fig. 4d, e). Therefore, the increased miR204 expression caused by IDH2 deficiency regulates CPT1a protein synthesis.
a Expected binding site of cpt1a mRNA and miR204 from Homo sapiens and Mus musculus. b Overexpression of the miR204 duplex decreased the protein level of WT CPT1a. Construction of Myc-tagged WT CPT1a-coding region and western blots. c Overexpression of the miR204 duplex did not affect the protein level of mutant cpt1a. Construction of Myc-tagged and CPT1a mutated plasmid coding region and western blots. d Western blot analysis of CPT1a in ND- or HFD-fed WT and KO mice. e Western blot analysis of CPT1a in IDH2-deficient and miR204 inhibitor-treated mouse hepatocytes. β-actin was used as the internal control. Data are means ± SEM of three independent experiments (n = 3). $p < 0.05 vs. KO-ND mice. *p < 0.05 vs. siCON. #p < 0.05 vs. KO-HFD mice transfected with a miR204 mimic (204-M; overexpression).
Inhibition of miR204 ameliorates fatty acid β-oxidation in the hepatocytes
Fatty acid β-oxidation is the process by which fatty acids are broken down to generate energy. The increased miR204 expression caused by IDH2 downregulation inhibits CPT1a, a key regulator of fatty acid β-oxidation in mitochondria (Fig. 5a). The expression levels of the upstream markers of fatty acid β-oxidation—AMP-activated protein kinase (AMPK) phosphorylation, peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC1α), and PPARα—were restored in miR204-I-injected WT and IDH2 KO mice (Fig. 5b). The mRNA levels of enzymes involved in fatty acid β-oxidation were confirmed in IDH2 KO mice and WT mice injected with miR204-I (Fig. 5c, d). CPT1a restoration rescued the levels of downstream molecules related to fatty acid β-oxidation, such as LCAD, MCAD, and UCP2. In vitro, we measured the fatty acid oxidation (FAO) activity by 18C-labeled unsaturated fatty acid oleate as the substrates with CPT1a inhibitor, Etomoxir (ETX) and the FAO activator FCCP as negative and positive controls, respectively. The FAO was decreased by IDH2 deficiency, however, inhibition of miR204 rescues FAO activity in IDH2-deficient hepatocytes (Supplementary Fig. 2). Therefore, miR204 blocks the degradation of fatty acids, leading to the accumulation of fat and cholesterol in the hepatocytes.
a Degradation of fatty acids. b Western blot analysis of the phosphorylation of AMPK, PGC1α, and PPARα in the liver of IDH2 KO and WT mice fed an HFD, and treated or not with an miR204 inhibitor. β-actin was used as the internal control. c, d Expression of genes related to cholesterol efflux and synthesis, fatty acid synthesis, and fatty acid β-oxidation in the liver of IDH2 KO and WT mice fed an HFD, and treated or not with a miR204 inhibitor, as determined by quantitative real-time PCR (normalized to GAPDH). Data are means ± SEM of three independent experiments (n = 3). *p < 0.05 vs. WT-HFD mice. #p < 0.05 vs. KO-HFD mice.
miR204 inhibition attenuates IDH2 deletion-induced NAFLD
To assess the role of miR204 in the development of fatty liver and obesity in IDH2 KO mice, we compared subcutaneous fat accumulation and blood parameters between WT and KO mice fed an ND or HFD for 12 weeks. miR204 inhibitors were injected subcutaneously (s.c.) using an osmotic pump for 4 weeks after feeding with the HFD for 8 weeks. Inhibition of miR204 significantly decreased the hepatic miR204 level (Fig. 6a). Inhibition of miR204 suppressed the increase in body weight of KO mice fed an ND or HFD (Fig. 6b, c). Inhibition of miR204 reduced the epididymal fat-pad weight in WT and KO mice fed an ND or HFD (Fig. 6d). Also, miR204 inhibition significantly rescued excessive lipid accumulation, as determined by H&E staining (Fig. 6e) and analysis of the serum levels of total cholesterol, triglyceride and LDL in WT and KO mice fed an HFD (Fig. 6f–h). Therefore, miR204 inhibition activates β-oxidation of fatty acid by restoring CPT1a expression in the liver.
a miR204 expression in liver tissues; RNU6 was used as the internal control. b Body weight of male mice injected with miR204-I and fed the ND (black-WT, blue-WT + 204-I, green-KO, red-KO + 204-I) or HFD (black-WT, blue-WT + 204-I, green-KO, red-KO + 204-I) at 1–12 weeks. c Photographs of KO mice treated or not with an miR204-I and fed an ND or HFD. d Epididymal fat mass. e Representative H&E-stained high-magnification images of mouse livers; scale bars, 50 μm. f–h Blood parameters. i Representative immunohistochemical images of CD68 in liver tissues of WT and KO-HFD-fed mice treated or not with miR204-I. Scale bars, 50 μm. j Western blot analysis of CD68 in liver tissues of two mice per group. k–m Expression of IL-1β (k), IL-6 (l), and TNF-α (m) by quantitative real-time PCR, normalized to GAPDH. Data are means ± SEM of three independent experiments (n = 3). *p < 0.05 vs. KO-ND mice. #p < 0.05 vs. WT-HFD mice. &p < 0.05 vs. KO-HFD mice.
The CD68 immunohistochemical signal intensity was decreased by miR204-I (Fig. 6i). Western blotting also showed that inhibition of miR204 rescued macrophage infiltration in the liver (Fig. 6j). Additionally, miR204-I-treated KO-HFD mice exhibited lower IL-1β, IL-6, and TNF-α mRNA levels than KO-HFD mice (Fig. 6k–m). These findings suggest that miR204 inhibition attenuates inflammation by inhibiting the synthesis of pro-inflammatory cytokines in KO mice.
Discussion
The prevalence of NAFLD has increased significantly in most developed countries over the past few decades. NAFLD involves low-level inflammation in insulin-targeted tissues such as muscle, fat, and liver25. Chronic inflammation increases the levels of chemokines and cytokines, inducing systemic embolic disorder in the liver26. Mitochondrial dysfunction promoted the development of metabolic diseases by modulating the level of active oxygen, the reducing power of oxygen radicals, and cellular energy production12. Maintaining mitochondrial homeostasis is a prerequisite for preventing metabolic diseases.
Deficiency of the mitochondrial protein IDH2 induces mitochondrial dysfunction, inflammation, and endothelium-dependent vasomotor function in endothelial cells9,10. In this study, IDH2 KO mice fed an ND or HFD exhibited severe fat accumulation, and elevated blood cholesterol and triglyceride levels, compared to WT mice (Fig. 1). Notably, IDH2 KO mice fed an HFD exhibited significantly increased levels of pro-inflammatory cytokines, and the expression of fatty acid and cholesterol efflux related genes (both of which regulate inflammation and lipid metabolism), compared to WT mice fed an HFD (Figs. 1 and 2).
IDH2 deletion increased obesity resistance and metabolic stress through brown adipose tissue whitening12,13. Therefore, we used IDH2 KO mice for NAFLD studies, assuming that IDH2 KO mice with HFD more accelerates the phenomenon of fat accumulation than WT mice with HFD. In addition, we investigated the direct mechanism of fat accumulation in the liver. IDH2 deficiency increased hepatic miR204 expression (Fig. 3). miR204 regulates cardiovascular diseases by modulating SIRT1 expression27,28, suppresses tumor-cell proliferation29 and regulates pancreatic β-cell proliferation30,31. We hypothesized that the increased miR204 expression caused by IDH2 downregulation regulates genes related to lipid metabolism. miRNAs inhibit gene expression by direct post-transcriptional gene silencing in the cytosol, achieved by binding to the 3′-UTR of the mRNA32 without altering target gene expression. However, miRNAs also inhibit target gene translation by directly binding the CDS24. The mRNA levels of MCAD, SCD1 and CPT1a were similar between ND and HFD IDH2 KO mice. The CDS of CPT1a was prioritized over other candidates to match the seed site of miR204 (Fig. 4). Also, there was no change in Myc expression in CPT1a mutant plasmid-transfected cells, indicating that miR204 inhibits CPT1a protein synthesis by matching the target sequence in the CDS of CPT1a (Fig. 4). A more important finding in this study was to clarify the correlation between liver fat accumulation due to IDH2 deficiency and miR204 in a high-fat diet (Pearson correlation coefficient) (Fig. 3d and Supplementary Fig. 1).
AMPK plays a central role in the regulation of energy homeostasis in the body, as well as fat metabolism33. Abnormalities in these energy sensors are related to the development of cardiovascular diseases, metabolic disorders, and cancer. Upon activation, AMPK suppresses the synthesis of fatty acids and cholesterol, and promotes the oxidation of fatty acids. HFD-fed IDH2 KO mice showed suppressed AMPK activation (phosphorylation) compared to HFD-fed WT mice (Fig. 5b). An important regulatory mechanism of AMPK is the inhibition via phosphorylation of acetyl-CoA carboxylase (ACC)34,35. ACC regulates lipid metabolism in multiple tissues, such as the liver and muscle, and ACC transforms acetyl-CoA into malonyl-CoA. Malonyl-CoA is present in the outer membrane of mitochondria and inhibits CPT1a. CPT1a plays an important role in β-oxidation in mitochondria. As expected, inhibition of miR204 attenuated fat accumulation and inflammation in this study, as indicated by the blood levels of several lipid markers and body and fat-pad weights in WT and KO mice (Fig. 6).
Our data demonstrate that the increased miR204 level caused by IDH2 deficiency modulates lipid metabolism by inhibiting CPT1a expression. Whitening of adipose tissue and mitochondrial dysfunction of adipocytes due to IDH2 deficiency are well known12. However, the effect of IDH2 deficiency on hepatocytes has not yet been reported. IDH2 deficiency causes citrate accumulation, which could be converted to Acetyl-CoA, and it can be a precursor to fatty acid synthesis in hepatocytes (Fig. 2b, c). In addition, there is no change in miR204 in WT-HFD mice compared to WT-ND (Fig. 3d). However, miR204 inhibition affects amelioration of NAFLD in WT-HFD (Figs. 4–6). As the deficiency of IDH2 and the effect of miR204 inhibitor affected not only hepatocytes but also cells of some organs (e.g., pancreatic beta cells, brown fat). We plan to enhance the originality of the disease mechanism through hepatocyte-specific IDH2 & miR204 double-deficient mice and study the association of miR204 / IDH2 in samples obtained from patients with non-alcoholic fatty liver disease. Despite the ever-increasing burden of NAFLD, it remains an unmet medical need with no approved pharmacologic treatment. Hence, strategies to regulate miR204 may have therapeutic potential for NAFLD and other liver diseases linked to mitochondrial dysfunction.
Methods
Cell culture and transfection
Mouse hepatocytes were purchased from the American Type Tissue Collection (Cat. CRL-2390; ATCC, Manassas, VA, USA) and cultured in F-12K medium (ATCC; Cat. #30-2004) containing 10% fetal bovine serum (Cat. #F-0500-A) (Atlas Biologicals, Fort Collins, CO, USA) and 1% antibiotic-antimycotic (Cat. #15240096; Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37 °C in 5% CO2 according to the manufacturer’s instructions. Hepatocytes were transfected with an siRNA targeting IDH2 (mouse siRNA sequence: sense, 5′-GCGACCAGUACAAGGCCACAGAUUU-3′; antisense, 5′-AAAUCUGUGGCCUUGUACUGGUCGC-3′) and a negative control siRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. For miR204 regulation, hepatocytes were transfected with miR204-I (5′-AGG ATG ACA AAG GGA-3′), a miR204 mimic sequence, or a scrambled nucleotide (5′-ACG TCT ATA CGC CCA-3′) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. For gene overexpression, hepatocytes were transfected with Myc-cpt1a (Cat. #100146; Addgene, Cambridge, MA, USA) full-length plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations. Transfected cells were incubated for 2 days at 37 °C in a 5% CO2 incubator.
Mouse studies
Experiments involving mice were approved by and followed the guidelines of the Institutional Animal Care and Use Committee at Chungnam National University Hospital (CNUH-019-A0016). The animals used were IDH2−/− germ-line KO mice, and their congenic background strain (C57BL/6J) served as the WT (IDH2+/+) control. Mice were maintained in a controlled environment (ambient temperature, 22–24 °C; humidity, 50–60%; 12 h light/dark cycle). IDH2 KO mice were a gift from Dr. Jeen-Woo Park (School of Life Sciences and Biotechnology, College of Natural Science, Kyungpook National University, South Korea). An HFD (60% of total calories come from fat, Teklad Custom Diet (Cat. TD.06414)-full composition: [Casein 265.0 g/kg, l-Cystine 4 g/kg, Maltodextrin 160 g/kg, Sucrose 90 g/kg, Lard 310 g/kg, Soybean Oil 30 g/kg, Cellulose 65.5 g/kg, Mineral Mix (AIN-93G-MX, 94046) 48 g/kg, Calcium Phosphate, dibasic 3.4 g/kg, Vitamin Mix (AIN-93-VX, 94047) 21.0 g/kg, Choline Bitartrate 3.0 g/kg]) was fed to WT male mice and IDH2 KO male mice (8 weeks old) for 12 weeks (Doo Yeol Biotech, Seoul, South Korea).
Immunoblotting
Cultured hepatocytes and liver tissues were harvested and lysed in 100 µl of RIPA buffer (1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 0.15 M NaCl, 0.05 M Tris-HCl; pH 8), containing 1× Halt protease inhibitor cocktail (Thermo Fisher Scientific) for 30 min on ice. Liver tissue samples were then homogenized for 3 min in RIPA buffer on ice. After clearing by centrifugation at 13,000 rpm for 10 min, the protein concentration was measured using a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific). Equal amounts of protein per well were resolved by 10–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose membrane. The membrane was washed with Tris-buffered saline (10 mM Tris, 150 mM NaCl) containing 0.1% Tween 20 (TBST) and blocked in TBST containing 5% bovine serum albumin Fraction V (Roche, Basel, Switzerland). Membranes were then incubated with the following antibodies: anti-IDH2 (ab129180), anti-CD68 (ab125212), anti-CPT1a (ab83862), anti-PPAR alpha (ab24509) (Abcam, Cambridge, UK); anti-PGC1α (sc-13067) (Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-phospho-AMPK (2531S), anti-AMPK (2532S) (Cell Signaling Technology, Danvers, MA, USA); and anti-β-actin (A-5441) (Sigma-Aldrich, St. Louis, MO, USA). Immunoblotting of 30 μg of whole-cell lysate or tissue homogenate was performed using appropriate primary and horseradish peroxidase (HRP)-conjugated secondary antibodies, and the chemiluminescent signal was developed using Super Signal West Pico or Femto Substrate (Pierce Biotechnology, Rockford, IL, USA). Values were normalized to β-actin as a loading control.
Real-time qPCR
Total RNA was isolated using TRIzol Reagent (Invitrogen) based on the acid guanidinium thiocyanate–phenol–chloroform method. The total RNA concentration was determined using a SmartSpec 3000 spectrophotometer (Bio-Rad, Hercules, CA, USA). Complementary DNA was prepared from total RNA using the Maxime RT Premix kit (iNtRON Biotechnology, Seongnam, Korea)). Quantitative real-time PCR was performed using the Prism7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) with the Super Script III Platinum SYBR GreenOne-Step qRT-PCR Kit (Invitrogen). The primers used for mouse ABCG1 were sense 5′-GAT TGG CTT CAG GAT GTC CAT GTT GGA A-3′ and antisense 5′-GTA TTT TTG CAA GGC TAC CAG TTA CAT TTG ACA A-3′; those for mouse CYP7A1 were sense 5′-CTG TGT TCA CTT TCT GAA GCC ATG-3′ and antisense 5′-CCC AGG CAT TGC TCT TTG AT-3′; those for mouse HMCGR and mouse SREBP-1c were sense 5′-AGG GGT AGG GCC AAC GGC CT-3′ and antisense 5′- CAA GCT GCC TGG GGA GCT GGT A-3′; those for mouse ACCa were sense 5′ -GGC CAG TGC TAT GCT GAG AT-3′ and antisense 5′ AGG GTC AAG TGC TGC TCC A-3′; those for mouse FAS were sense 5′-CTG CGG AAA CTT CAG GAA ATG-3′ and antisense 5′-GGT TCG GAA TGC TAT CCA GG-3′; those for mouse PPARγ were sense 5′-CAA GAC TAC CCT TTA CTG AA-3′ and antisense 5′-CTA CTT TGA TCG CAC TTT GGT-3′; those for mouse SCD1 were sense 5′-CTT CTT GCG ATA CAC TCT GG-3′ and antisense 5′-TGA ATG TTC TTG TCG TAG GG-3′; those for mouse ACOX-1 were sense 5′-GAA TTT GGC ATC GCA GAC CC-3′ and antisense 5′-GAT CTC CAG ATT CCA GGC CG-3′; those for mouse CPT1a were sense 5′-TCT TGC AGT CGA CTC ACC TT-3′ and antisense 5′-TCC ACA GGA CAC ATA GTC AGG-3′; those for mouse LCAD were sense 5′-AAG GAT TTA TTA AGG GCA AGA AGC-3′ and antisense 5′-GGA AGC GGA GGC GGA GTC-3′; those for mouse MCAD were sense 5′-GAG CCT GGG AAC TCG GCT TGA-3′ and antisense 5′-GCC AAG GCC ACC GCA ACT TT-3′; those for mouse PDK4 were sense 5′-TGT GAT GTG GTA GCA GTA GTC-3′ and antisense 5′-GTG GTG AAG GTG TGA AGG A-3′; those for mouse PPARα were sense 5′-CAA GGC CTC AGG GTA CCA CT-3′ and antisense 5-‘TTG CAG CTC CGA TCA CAC TT-3′; those for mouse UCP2 were sense 5′-GTT CCT CTG TCT CGT CTT GC-3′ and antisense 5′-GGC CTT GAA ACC AAC CA-3′; and those for mouse GAPDH were sense 5′-GGT GAA GGT CGG TGT GAA CG-3′ and antisense 5′-CCC GTA GGG CGA TTA CAG TC-3′ (internal control). Dissociation curves were generated to check for primer dimers. Fold-changes in gene expression were calculated by the 2–ΔΔCt method.
miRNA isolation and qPCR
Total RNA was isolated using TRIzol Reagent (Invitrogen) based on the acid guanidinium thiocyanate–phenol–chloroform method. The total RNA concentration was determined using a SmartSpec 3000 spectrophotometer (Bio-Rad). Complementary DNA was prepared from total RNA using the miScript II RT Kit (Cat. #218161; Qiagen, Hilden, Germany). Quantitative real-time PCR was performed using the Prism7000 Sequence Detection System (Applied Biosystems) with the miScript SYBR Green PCR Kit (Cat. #218073; Qiagen). The primer used for mmu-miR204 was sense 5′-CGC TTC CCT TTG TCA TCC TA-3′; mmu-miR125-2-3p was sense, 5′-GTCAGGCTCTTGGGACAAAA-3′; mmu-miR148a-5p was sense, 5′-GTTCTGAGACACTCCGACTAAAA-3′; mmu-miR188-5p was sense, 5′-CATCCCTTGCATGGTGGAG-3′; mmu-miR451a was sense, 5′-CGAAAACCGTTACCATTACTGA-3′; mmu-miR700-5p was sense, 5′-GCTTACAATCTAGCTGGGAAAAA-3′; mmu-miR714 was sense, 5′-CCCAGGGAGAGACGTAAGAA-3′; mmu-miR802-3p was sense, 5′-CGCAGTAACAAAGATTCATCCTTGTAAAA-3′ and the antisense primer was a universal primer included in the SYBR Green PCR kit. The primer used for RNU6 was sense 5′-GCA AAT TCG TGA AGC GTT CC-3′ and the antisense primer was a universal primer included in the SYBR Green PCR Kit (internal control). Dissociation curves were generated to check for primer dimers. Fold-changes in gene expression were calculated by the 2−ΔΔCt method.
Histological analysis
After washing with phosphate-buffered saline, tumor tissues were fixed in 4% (w/v) paraformaldehyde and embedded in paraffin. Paraffin sections were deparaffinized and rehydrated according to standard protocols and stained with H&E. For immunohistochemistry, tumor tissues were stained with an anti-CD68 primary antibody (diluted 1:100; Cat. #ab125212; Abcam) overnight at 4 °C. Next, an HRP-conjugated anti-rabbit IgG was added for 60 min at room temperature. Color was developed by incubation for 5 s with 3,3′-diaminobenzidine (DAB). Sections were counterstained with hematoxylin and examined under a microscope (Motic, Richmond, BC, Canada) at 100× magnification.
In vivo inhibition of miR204
An inhibitor of miR204 (5′-AGG ATG ACA AAG GGA-3′; miR204-I) (Bioneer, Daejeon, South Korea) was systemically infused into mice for 4 weeks (0.5 mg/kg/day) using an ALZET Osmotic Pump (Cat. 1004) implanted subcutaneously.
Citrate assay
Livers were extracted and homogenized in citrate assay buffer. The citrate concentration was determined using a Citrate Colorimetric/Fluorometric Assay Kit (Cat. #K655-100; Biovision, Milpitas, CA, USA) according to the manufacturer’s instructions.
Acetyl-CoA assay
Livers were extracted and homogenized in acetyl-CoA assay buffer. The acetyl-CoA concentration was determined using a PicoProbe™ Acetyl-CoA Colorimetric/Fluorometric Assay Kit (Cat. #K317; Biovision) according to the manufacturer’s instructions.
Blood analysis
Blood parameters (glucose, cholesterol, triglycerides, high-density lipoprotein, and LDL) were analyzed using a Samsung LABGEOPT Lipid Test 5 Kit with a Samsung LABGEO PT10 Analyzer according to the manufacturer’s instructions.
Fatty acid in vitro treatment and Oil Red O staining
After 24 h transfection, these cells treated fatty acid (1 mM: Oleic acid 0.66 mM and Palmitic acid 0.33 mM) with cell growth media and then incubation for 24 h. Cells were then washed three times with ice PBS and fixed in 4% Paraformaldehyde 30 min at room temperature. After fixation, cells were washed three times with PBS and stained Oil red O solution (working solution, 0.5 g Oil Red O powder dissolved in 60% isopropanol) for 30 min at room temperature. Cells were washed again with PBS to remove unbound staining. To quantify Oil Red O content levels, dimethyl sulfoxide was added to each sample; after shaking at room temperature for 5 min and then were analyzed using a microscope.
Fatty acid oxidation (FAO) assay
FAO assay was following the manufacturer’s instruction of Fatty Oxidation Assay Kit (Cat. ab217602, Abcam). Hepatocyte 1 × 104 cells were seeded per well in 96 well plates and growing overnight. The cells transfected with siCON, siIDH2 and siIDH2 + miR204 inhibitor three wells per condition. The wells without cells were used as signal control. After 24 h, the cells rinsed twice with 100 μl pre-warmed FA-Free media followed by adding 90 μl pre-warmed FA measurement Medium. A total of 85 μl of FA-Free measurement medium was added to the wells, and 5 μl of BSA control were included as the FA-Free control. All wells except the signal control had 10 μl extracellular oxygen consumption reagent added from Extracellular Oxygen Consumption Assay Kit (Cat. Ab197243, Abcam). FCCP (0.625 μM, FAO activator) and Etomoxir (40 μM, CPT1a inhibitor, FAO inhibitor) added were included as signal control. And then the cells were sealed with 100 μl pre-warmed mineral oil, and the FAO signal were collected at 5 min intervals for 90 min at Ex/Em = 380/650 nm. The proteins were normalized by protein concentration using the BCA assay.
Statistical analysis
Statistical analysis was performed using Prism 8 software (GraphPad Software Inc., La Jolla, CA, USA). Data are means ± standard error of the mean (SEM). Differences between two groups were evaluated using t-tests. For multiple comparisons, one-way analysis of variance (ANOVA) was performed followed by a Tukey’s multiple comparison test. p-values < 0.05 were considered indicative of statistical significance. Data are representative of at least three independent experiments.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The datasets supporting the conclusions of this article are available from the corresponding author upon reasonable request.Original data related to this work: Kim, Seonhee; Kim, Cuk-Seong (2022), Western blot quantification, Dryad, Dataset, https://doi.org/10.5061/dryad.crjdfn36h. Unprocessed data Sharing Link: https://datadryad.org/stash/share/ClKxEuHhpvKsX722hq4doBx7qNqneRCiX4ZhD5y4J70. Uncropped and unedited blot/gel images are listed in Supplementary Fig. 3.
References
Noh, M. R., Kong, M. J., Han, S. J., Kim, J. I. & Park, K. M. Isocitrate dehydrogenase 2 deficiency aggravates prolonged high-fat diet intake-induced hypertension. Redox Biol. 34, 101548 (2020).
Huang, P. L. A comprehensive definition for metabolic syndrome. Dis. Model Mech. 2, 231–237 (2009).
Younossi, Z. M. et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64, 73–84 (2016).
Wang, F. et al. Activated natural killer cell promotes nonalcoholic steatohepatitis through mediating JAK/STAT pathway. Cell Mol. Gastroenterol. Hepatol. 13, 257–274 (2021).
Furukawa, S. et al. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Invest. 114, 1752–1761 (2004).
Leskov, I. et al. Nicotinamide nucleotide transhydrogenase activity impacts mitochondrial redox balance and the development of hypertension in mice. J. Am. Soc. Hypertens. 11, 110–121 (2017).
Bhatti, J. S., Bhatti, G. K. & Reddy, P. H. Mitochondrial dysfunction and oxidative stress in metabolic disorders—a step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 1066–1077 (2017).
McInnes, J. Mitochondrial-associated metabolic disorders: foundations, pathologies and recent progress. Nutr. Metab. (Lond.) 10, 63 (2013).
Park, J. B. et al. IDH2 deficiency impairs mitochondrial function in endothelial cells and endothelium-dependent vasomotor function. Free Radic. Biol. Med. 94, 36–46 (2016).
Choi, S. J. et al. Isocitrate dehydrogenase 2 deficiency induces endothelial inflammation via p66sh-mediated mitochondrial oxidative stress. Biochem. Biophys. Res. Commun. 503, 1805–1811 (2018).
Du, J. et al. Reductive carboxylation is a major metabolic pathway in the retinal pigment epithelium. Proc. Natl Acad. Sci. USA 113, 14710–14715 (2016).
Lee, J. H. et al. Isocitrate dehydrogenase 2 protects mice from high-fat diet-induced metabolic stress by limiting oxidative damage to the mitochondria from brown adipose tissue. Exp. Mol. Med. 52, 238–252 (2020).
Lee, S. J., Kim, S. H., Park, K. M., Lee, J. H. & Park, J. W. Increased obesity resistance and insulin sensitivity in mice lacking the isocitrate dehydrogenase 2 gene. Free Radic. Biol. Med. 99, 179–188 (2016).
Sun, W. et al. Hepatic CPT1A facilitates liver-adipose cross-talk via induction of FGF21 in mice. Diabetes https://doi.org/10.2337/db21-0363 (2021).
Krutzfeldt, J. & Stoffel, M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 4, 9–12 (2006).
Trajkovski, M. et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474, 649–653 (2011).
Klein, D. et al. MicroRNA expression in alpha and beta cells of human pancreatic islets. PLoS ONE 8, e55064 (2013).
Bordone, L. et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 4, e31 (2006).
Marzinotto, I. et al. miR-204 is associated with an endocrine phenotype in human pancreatic islets but does not regulate the insulin mRNA through MAFA. Sci. Rep. 7, 14051 (2017).
Tacke, F. Functional role of intrahepatic monocyte subsets for the progression of liver inflammation and liver fibrosis in vivo. Fibrogenes. Tissue Repair 5, S27 (2012).
Arango Duque, G. & Descoteaux, A. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol. 5, 491 (2014).
Cheung, O. & Sanyal, A. J. Role of microRNAs in non-alcoholic steatohepatitis. Curr. Pharm. Des. 16, 1952–1957 (2010).
Rottiers, V. & Naar, A. M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 13, 239–250 (2012).
Lu, X., Wei, Y. & Liu, F. Direct regulation of p53 by miR-142a-3p mediates the survival of hematopoietic stem and progenitor cells in zebrafish. Cell Discov. 1, 15027 (2015).
Xu, M. et al. Functional loss of inactive rhomboid-like protein 2 mitigates obesity by suppressing pro-inflammatory macrophage activation-triggered adipose inflammation. Mol. Metab. 34, 112–123 (2020).
Ellulu, M. S., Patimah, I., Khaza’ai, H., Rahmat, A. & Abed, Y. Obesity and inflammation: the linking mechanism and the complications. Arch. Med. Sci. 13, 851–863 (2017).
Vikram, A. et al. Vascular microRNA-204 is remotely governed by the microbiome and impairs endothelium-dependent vasorelaxation by downregulating Sirtuin1. Nat. Commun. 7, 12565 (2016).
Kassan, M. et al. Corrigendum: Sirtuin1 protects endothelial Caveolin-1 expression and preserves endothelial function via suppressing miR-204 and endoplasmic reticulum stress. Sci. Rep. 7, 46123 (2017).
Liang, Y., Tian, W. Y., Huang, J. J., Gao, L. X. & Fan, X. H. MicroRNA-204 plays a role as a tumor suppressor in Newcastle disease virus-induced oncolysis in lung cancer A549 cells. Oncol. Lett. 21, 482 (2021).
Gaddam, R. R. et al. Microbiota-governed microRNA-204 impairs endothelial function and blood pressure decline during inactivity in db/db mice. Sci. Rep. 10, 10065 (2020).
Gaddam, R. R. et al. Genetic deletion of miR-204 improves glycemic control despite obesity in db/db mice. Biochem. Biophys. Res. Commun. 532, 167–172 (2020).
Fan, L. et al. miR-552-3p modulates transcriptional activities of FXR and LXR to ameliorate hepatic glycolipid metabolism disorder. J. Hepatol. 74, 8–19 (2021).
Wang, Q., Liu, S., Zhai, A., Zhang, B. & Tian, G. AMPK-mediated regulation of lipid metabolism by phosphorylation. Biol. Pharm. Bull. 41, 985–993 (2018).
Lee, M. et al. Phosphorylation of acetyl-CoA carboxylase by AMPK reduces renal fibrosis and is essential for the anti-fibrotic effect of metformin. J. Am. Soc. Nephrol. 29, 2326–2336 (2018).
Galic, S. et al. AMPK signaling to acetyl-CoA carboxylase is required for fasting- and cold-induced appetite but not thermogenesis. Elife 7, https://doi.org/10.7554/eLife.32656 (2018).
Acknowledgements
The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see: http://www.textcheck.com/certificate/cC0qtH. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2014R1A6A1029617 and NRF-2019R1I1A3A01059497) and by the research fund of Chungnam National University. Kaikobad Irani was supported by funding from R01HL147545, R01HL115955, VA Merit Award I01BX002940, and ONR N0001142012147.
Author information
Authors and Affiliations
Contributions
S.K. and C.S.K. initiated and designed the study. S.K. performed most of the experiments. S.K. and C.S.K. wrote the manuscript. S.P, I.L., and H.N. performed some part of the experiments. I.L., Y.R.K., S.P., and S.C. performed data curation. C.S.K., B.H.J., and K.I. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary handling editors: Christopher Hine and Manuel Breuer.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, S., Lee, I., Piao, S. et al. miR204 potentially promotes non-alcoholic fatty liver disease by inhibition of cpt1a in mouse hepatocytes. Commun Biol 5, 1002 (2022). https://doi.org/10.1038/s42003-022-03945-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-022-03945-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.