Abstract
TDP-43 (TAR DNA-binding protein 43) aggregation and redistribution are recognised as a hallmark of amyotrophic lateral sclerosis and frontotemporal dementia. As TDP-43 inclusions have recently been described in the muscle of inclusion body myositis patients, this highlights the need to understand the role of TDP-43 beyond the central nervous system. Using RNA-seq, we directly compare TDP-43-mediated RNA processing in muscle (C2C12) and neuronal (NSC34) mouse cells. TDP-43 displays a cell-type-characteristic behaviour targeting unique transcripts in each cell-type, which is due to characteristic expression of RNA-binding proteins, that influence TDP-43’s performance and define cell-type specific splicing. Among splicing events commonly dysregulated in both cell lines, we identify some that are TDP-43-dependent also in human cells. Inclusion levels of these alternative exons are altered in tissues of patients suffering from FTLD and IBM. We therefore propose that TDP-43 dysfunction contributes to disease development either in a common or a tissue-specific manner.
Similar content being viewed by others
Introduction
TDP-43 (TAR DNA-binding protein 43), a protein encoded by the TARDBP gene, is a ubiquitously expressed member of heterogeneous nuclear ribonucleoprotein (hnRNP) family able to bind DNA and RNA that participates in various steps of mRNA metabolism, including transcription, pre-mRNA splicing, miRNA generation, regulation of mRNA stability, nucleo-cytoplasmic transport and translation1,2,3,4. TDP-43 was initially described as the major component of cytoplasmic inclusions formed in motor neurons of patients suffering from amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTLD) despite the fact that mutations in TARDBP gene only account for a small subset of those cases5,6,7. However, TDP-43 aggregates have also been found in skeletal muscles of patients with inclusion body myositis (IBM)8,9, oculopharyngeal muscular dystrophy (OPMD)10 and limb-girdle muscular dystrophy type 2a (LGMD2a)11 suggesting that TDP-43 aggregation may play a prominent pathological role also in muscle tissue. Accordingly, TDP-43 myogranules have been shown to provide essential functions during skeletal muscle development and regeneration, both in mouse and human12. Despite ubiquitous expression of TDP-43, however, most studies investigating this protein have focused on its role in the central nervous system. Nonetheless, given its importance of TDP-43, both in muscle development and potentially in the pathogenesis of numerous myopathies, we have now systematically investigated functions elicited by TDP-43 in muscle (C2C12) and neuronal (NSC34) mouse cells in parallel.
Performing such a comparison is particularly interesting as these two cell environments display tissue-characteristic features, like for example: distinct post-translational modifications (PTMs) and cleavage products of TDP-43 described in muscles and neurons13, muscle-characteristic localization of TDP-43 in space and time12, cell-type-specific milieu of TDP-43 binding partners14, and differential expression of RNA-binding proteins (RBPs) controlling common mRNA targets15,16. It is important to note that all these differences occur in a context of highly variable transcriptome between tissues including non-coding transcripts17,18,19. Therefore, TDP-43 might likely elicit tissue characteristic functions by targeting unique subsets of transcripts, which encode proteins participating in tissue-specific cellular pathways and provide crucial structural and functional features of a cell. The consequences of TDP-43 dysfunction in muscles could thus possibly differ from those that have so far been described in the central nervous tissue20,21.
In the last decade, high throughput methodologies have shifted the focus from characterization of individual events towards less biased global approaches, setting the ground for a systematic comparison of TDP-43 targeted RNAs across tissues and conditions. However, the overlap of TDP-43-controlled events identified by earlier studies is rather poor. It probably reflects the variation in technical approaches (microarrays, RNA-seq, CLIP-seq) and models employed in those studies: mouse brain20, human post-mortem brain samples21,22, human neuroblastoma cell line SH-SY5Y21,23, HEK-29324,25, Hela26. A clearer understanding of the extent to which TDP-43-mediated events are conserved between mouse and human is still lacking, yet it is a crucial point that should be addressed in future as it will allow better comparisons of human and mouse models of disease.
To finally address this issue in a systematic manner, we have identified subsets of unique cell-type-specific mRNA targets, as well as commonly regulated mRNAs, the tight regulation of which might underlie functions crucial for cell survival. More specifically, we have further explored splicing events that commonly occur in C2C12 and NSC34 cells and are additionally conserved in humans. We finally show that inclusion of common mouse-human TDP-43-regulated alternative exons is indeed altered in skeletal muscles of IBM patients and different brain regions of ALS and FTLD patients with reported TDP-43 pathology.
Results
TDP-43 expression is similar in C2C12 and NSC34 cells
To start comparing the functions of TDP-43 in cells of muscular and neuronal origin, we used the most commonly employed mouse cell lines representing skeletal muscle (C2C12) and motor neurons (NSC34). They have been previously used to study TDP-43-associated neurodegeneration as well as the role of TDP-43 in muscle development12,27,28,29. We first assessed protein levels of endogenous TDP-43 in untreated cells (Fig. 1a). Although in mature mouse tissues TDP-43 expression was reported to be higher in the brain compared to quadriceps muscle30, we noted no difference in the amount of total TDP-43 protein between undifferentiated C2C12 and NSC34 cells (Fig. 1a), nor in the expression of TDP-43 at the RNA level of siLUC-transfected cells (Fig. 1b, Supplementary Data 1).
a Western blot shows similar expression of endogenous TDP-43 in C2C12 and NSC34 cells. The amount of TDP-43 was normalized to the sum of peak intensities of 3 loading controls (tubulin, HSP70 and P84) (n = 3 replicates per group). b Expression levels of Tardbp in TDP-43-silenced C2C12 and NSC34 and corresponding controls assessed by RNA-sew plotted as log10-transformed FPKM values show TDP-43 was depleted (on the mRNA level) to the same extent in both cell lines (n = 3 replicates per group). padj = 1.6·10−18 for C2C12 and padj = 2.3·10−72 for NSC34. p-values were generated using Wald test and Benjamini–Hochberg multiple testing correction74. c Western blot shows reduction of TDP-43 in C2C12 and NSC34 cells upon siTDP transfection. siLUC-transfected cells were used as a control. TDP-43 expression was normalized against GAPDH (n = 3 replicates per group). d TDP-43 depletion led to altered splicing of Poldip3 and Sort1. Semi-quantitative RT-PCRs conducted in TDP-43-silenced samples and corresponding controls are shown along with the quantification of splicing changes (% of alternative exon inclusion). The number of the alternative exon is given below (see the exact transcript numbers in Supplementary Table 1, n = 3 replicates per group). e PCA plot visualizes distances between siLUC- and siTDP-transfected C2C12 and NSC34 cells based on FPKM of all genes obtained by RNA-seq (left). Variation in the PC2 is explained by the presence/absence of TDP-43 (right).
TDP-43 was silenced to a similar extent in both cell lines (Fig. 1b, Supplementary Data 1), and reduction of the protein was confirmed by western blot (Fig. 1c). TDP-43 loss functionally reflected in altered splicing of the two well characterized target transcripts Poldip3 and Sort1 (Fig. 1d)23,26,31,32. To explore transcriptome-wide effects of TDP-43 downregulation, we then performed RNA-seq analysis on polyadenylated mRNA extracted from TDP-43 depleted cells. Both cell lines displayed a characteristic transcriptome as revealed by principal component analysis (PC1), whereas the effect of TDP-43 knockdown explained a smaller portion of the variation between samples (PC2) (Fig. 1e, Supplementary Data 1). This result suggests that TDP-43 silencing promotes alterations in overall mRNA abundance in C2C12 and NSC34 based on the tissue-characteristic transcriptome.
mRNAs dysregulation following TDP-43 reduction in C2C12 and NSC34 cells is cell-type specific
Tissues substantially vary with regards to abundance of transcripts encoded by individual genes and splice isoforms they express, and these differences underlie specific biological characteristics and functions. To examine the effect of TDP-43 loss on expression levels (differential gene expression, DEG) in two cell lines, we separately normalized reads of C2C12 and NSC34 datasets and obtained 4019 transcripts, expression levels of which were subject to TDP-43 regulation. At padj < 0.05, we detected a very similar number of DEG in C2C12 and NSC34 (2325 and 2324, respectively), with 630 (15.7%) transcripts being commonly dysregulated in both cell lines (Fig. 2a, Supplementary Data 2). Surprisingly enough, the small overlap could not be explained by the fact that some genes are expressed in a cell-type-specific manner (i.e., muscle-characteristic genes are not transcribed in neuronal cells and vice versa), as the overlap between TDP-43 targets remained small (19.3%) even if we only considered genes expressed in both cell lines (FPKM in both cell lines > 0.5) (Fig. 2b, Supplementary Data 1 and 2). However, our data indicated that TDP-43 targets regulated in a cell-type-specific fashion are highly expressed in one cell type but not in the other. On average, C2C12-specific TDP-43-regulated mRNAs show higher expression in C2C12 than in NSC34 cells and vice versa (Supplementary Fig. 1a).
a Venn diagram shows the number of TDP-43-regulated transcripts identified in C2C12 and NSC34 cells exclusively (1695 and 1694, respectively), along with those that are commonly regulated by TDP-43 in both cell types (630). Transcripts with padj < 0.05 were considered as differentially expressed irrespective of their log2fold change. b Venn diagram shows the overlap (599 transcripts, 19.3%) of TDP-43-regulated DEG identified in C2C12 and NSC34 cell line (as in a, Supplementary Data 2), considering only transcripts expressed in both cell lines (FPKM in both cell lines > 0.5). Transcripts with padj < 0.05 were considered as differentially expressed irrespective of their log2-fold change. c Expression changes of common targets (a, Supplementary Data 2, 630) are plotted by their log2-fold change values in C2C12 and NSC34 (Spearman’s ρ = 0.77, p-value < 2.2 · 10−16). Grey line represents y = x and the blue line represents the fitted regression. d TDP-43-mediated expression changes in C2C12 and NSC34 represented as volcano plots. C2C12- and NSC34-specific targets are shown in red and blue, respectively, while common targets are plotted as grey dots. Vertical lines indicate fold changes of 0.7 (30% increase) and 1.3 (30% decrease). Best hits are labelled with gene names. e Venn diagram shows the number of cell-type-specific and overlapping GO terms enriched by DEG identified in C2C12 or NSC34 cells. GO terms (category: biological process) were grouped based on their names as those implying muscle- (red) or neuron-related features (blue). f Representative GO terms (category: biological process) commonly enriched by DEG in C2C12 and NSC34 cells suggesting pathological abnormalities described in neurodegenerative and myodegenerative disease (hand curated).
It has previously been proposed that TDP-43 binding is needed to sustain pre-mRNA levels and that mRNA downregulation would be a direct consequence of TDP-43 loss, while mRNA upregulation was explained by indirect effects20. In our datasets (Fig. 2a, Supplementary Data 2), the number of downregulated genes slightly outnumbered genes that were upregulated following TDP-43 depletion (Supplementary Fig. 1b); however, the overlap was very similar, irrespective the direction of the change (14.0% and 15.0% for upregulated and downregulated transcripts, respectively). Comparing the extent of expression changes of commonly regulated transcripts (630) induced by TDP-43 reduction, we saw a positive correlation (ρ = 0.77, p-value < 0.001) between two cell lines, with a trend towards larger alterations in C2C12 (Fig. 2c, Supplementary Data 2). Of note, there were few mRNAs whose expression was altered in the opposite direction in two cell lines, indicating that TDP-43 loss can elicit contrary effects depending on the cellular environment. Looking at individual target transcripts (Fig. 2d, Supplementary Data 2, Supplementary Fig. 1c, d), we hypothesized that the biggest expression changes induced by TDP-43 loss occurred in highly abundant transcripts. However, plotting the size of the change (log2 fold change) against background expression levels (FPKM in siLUC-transfected cells) of all DEG revealed that there is in fact no correlation between the two (Supplementary Fig. 1e).
Taken together, these results support the idea that unique sets of transcripts controlled by TDP-43 in each cell type can only partially be explained by variable expression levels of cell-type-characteristic mRNAs across tissues. Factors other than expression levels as such thus influence TDP-43 function that seems to be cell-type-specific. At the sequence level, in fact, TDP-43-regulated mRNAs detected in C2C12 or NSC34 appear to be equally well conserved across species (Supplementary Fig. 1f).
Commonly enriched processes implicated in neurodegenerative and myodegenerative disease
In mouse brain, TDP-43 has been shown crucial for maintenance of mRNAs that encode proteins involved in synaptic activity20. To elucidate which cellular processes might be controlled by TDP-43 in cells of muscle and neuronal origin, we conducted enrichment analysis of genes differentially expressed in C2C12 (2325) and NSC34 (2324) (Fig. 2e, Supplementary Data 5). Among C2C12 enriched GO terms, we found those directly associated with muscle characteristic features like striated muscle development or muscle cell migration, in line with results highlighting the importance of TDP-43 in skeletal muscle formation and regeneration12,29. On the other hand, a great portion of neuronal processes like vesicle-mediated transport in synapse or regulation of postsynaptic membrane neurotransmitters appeared to be affected by TDP-43 loss in NSC34 cells.
While the percentage of overlapping DEG was only 15.7%, by GO categories, almost a third of all biological processes (28%) enriched in C2C12 or NSC34 DEG (Fig. 2e, Supplementary Data 5) was commonly dysregulated upon TDP-43 depletion in both cell lines. Given that currently proposed picture of pathological processes implicated in myopathies bears several similarities with neurodegenerative disease9,33,34, we investigated commonly enriched GO terms to see if any of them could detect abnormalities previously described in the above-mentioned diseases. Significant GO terms enriched by DEG in both C2C12 and NSC34 (Fig. 2f, Supplementary Data 5) suggest that some common TDP-43-mediated mechanisms might contribute to development of TDP-43-proteinopathies in both muscle and neuronal tissues. Pathomechanisms include aberrant protein accumulation (i.e., of ubiquitin, amyloid β, α-synuclein, phosphorylated τ and TDP-43), post-translational modifications of deposited proteins (phosphorylation, ubiquitination, acetylation, sumoylation), defects in protein disposal (26 S proteasome and autophagy) and mitochondrial abnormalities. However, while there was a greater overlap between biological response to TDP-43 depletion (GO: biological process), the specific differentially expressed transcripts in common terms were remarkably different between C2C12 and NSC34 (Supplementary Fig. 1g). This implies that TDP-43 can influence similar biological processes in both muscles and neurons, but it does so by mediating expression levels of genes encoding for distinct proteins that participate in those pathways.
TDP-43-mediated splicing is more pronounced in NSC34 cells
Along with mRNA depletion, aberrant pre-mRNA splicing has been described to contribute to neuronal vulnerability as a consequence of pathologic TDP-43 behaviour20,21,35. Yet, little is understood about how TDP-43 dysfunction affects pre-mRNA splicing in tissues beyond the central nervous system. In this work, we systematically compared alternative splicing (AS) alterations following TDP-43 reduction in C2C12 and NSC34 cells. As expected, a considerably lower number of splicing events was detected in C2C12 than in NSC34 cells (730 and 1270, respectively) using rMATS at FDR of 0.01 (Fig. 3a, Supplementary Data 3), which held true for events of any classical AS category (i.e., SE, MXE, RI, A3'SS, A5'SS) (Fig. 3b, Supplementary Data 3). Neuronal and muscle targets did not vary with regard to event type proportion (Supplementary Fig. 2a); length of cassette exons (Supplementary Fig. 2b); the ratio between inclusion/exclusion events (Supplementary Fig. 2c); or percentage of frame-conserving events (Supplementary Fig. 2d). Interestingly enough, alternative sequences regulated by TDP-43 in the neuronal cell line seem to be more conserved across species than TDP-43-regulated sequences in muscle cell line (Supplementary Fig. 2e). This holds true particularly for cassette exons (Supplementary Fig. 2f), which represent the most frequent event type detected by our pipeline (Supplementary Fig. 2a).
a Venn diagram shows the total number of AS events (detected by rMATS at FDR < 0.01) induced by TDP-43 depletion in C2C12 and NSC34 specifically (630 and 1170, respectively), together with those commonly detected in both cell lines (100). b The number of annotated AS events (Fig. 3a, Supplementary Data 3) visualized by event type. SE - exon skipping, MXE - mutually exclusive exons, RI - intron retention, A3'SS and A5'SS - alternative 3' or 5' splice site. The percentage of overlapping AS events is reported on the plot. c Venn diagram shows the number of AS events (junctions) detected by MAJIQ (∆PSI > 0.2, FDR < 0.1, Supplementary Data 4). d Venn diagram showing the number of AS events detected by rMATS (Fig. 3 a, Supplementary Data 3) and MAJIQ (Fig. 3b, Supplementary Data 3), that were additionally validated using Interactive Genome Viewer.
The observation that TDP-43 regulates more events in NSC34 cells might reflect the importance of alternative splicing as a regulatory mechanism in neurons and support the existence of a distinct splicing programme in neuronal tissues, as already suggested by others14,36,37. Moreover, very few AS events detected by rMATS (on average 5.2%) appear to be commonly regulated by TDP-43 in both cell types, with the percentage of overlapping AS events being small (5.8%) even when we only considered AS in transcripts commonly expressed in both cell lines (FPKM > 0.5) (Supplementary Fig. 2g).
Even though Jeong et al.30 have previously reported that TDP-43’s repression of cryptic exons is tissue-specific, the percentage of overlapping regular (annotated) AS events seemed unexpectedly low. We therefore conducted an independent analysis using an alternative splicing tool—MAJIQ38, (∆PSI > 0.2, FDR < 0.1) (Fig. 3c, Supplementary Data 4). Like with rMATS, a higher number of TDP-43-dependent splicing events was detected in NSC34 cells (305 and 394 in C2C12 and NSC34, respectively). MAJIQ and rMATS quantify in separate ways (junctions or events, respectively)39; thus, comparable results are only produced when the same pipeline is applied. The proportion of commonly regulated annotated splicing events obtained by MAJIQ relative to rMATS (15.5% and 5.2%, respectively) clearly points out to the different performance of two event-based methods on the same dataset39.
To answer the question how cell-type-specific the activity of TDP-43 really is, we, as our final approach, estimated the overlap of TDP-43-controlled splicing between C2C12 and NSC34 cell line on a smaller subset of AS events, all of which were detected by both splicing tools (even if at the expense of false negatives). Moreover, we visually validated every event found by splicing tools using Interactive Genome Viewer and subsequently excluded those whose changes could not be confirmed (Fig. 3d, Supplementary Data 3 and 4).
Amongst these events, 45% of TDP-43-regulated splicing activity seemed to be shared between NSC34 and C2C12, and we firmly believe this provides a better estimate of TDP-43’s cell-type-specificity.
Alternatively spliced TDP-43 targets are implicated in neuronal functions and DNA-related processes
We further employed GO analysis to see whether genes with TDP-43-regulated splicing identified in C2C12 and NSC34 form interconnected networks and if TDP-43 can, by mediating AS, influence particular biological processes in each cell type. Since the number of C2C12 AS genes entering GO analysis (578) was considerably lower than that of NSC34 genes (1018), the analysis resulted in fewer GO terms found to be enriched in C2C12 compared to many in NSC34 (23 and 203, respectively) (Fig. 4a, Supplementary Data 3 and 5). As expected, GO terms enriched in NSC34 cells exclusively suggest that in these cells, alternatively spliced mRNA predominantly encode for proteins implicated in processes taking place in the nervous system (e.g., axonogenesis, regulation of neuron differentiation) (Fig. 4b, Supplementary Data 5). This is in line with earlier studies, which demonstrated that in human neuroblastoma cells SH-SY5Y TDP-43-dependent splice isoforms encode for proteins regulating neuronal development and those involved in neurodegenerative disease21.
a Venn diagrams show the number of alternatively spliced transcripts (as detected by rMATS at FDR < 0.01 (Fig. 3a, Supplementary Data 3)) in C2C12 and NSC34 cells together with GO terms (category: biological process, padj < 0.05) enriched in AS genes detected in each cell line. b GO terms uniquely enriched in NSC34 (198) imply on deregulation of neuronal processes, mRNA metabolism and DNA biology in NSC34 cells (representative GO terms are shown on the plot). c GO terms uniquely enriched in C2C12 (18) suggest involvement of TDP-43-regulated AS genes in DNA-modifying processes (representative GO terms are shown on the plot).
On the other hand, GO terms (56%) enriched in C2C12 cells exclusively (Fig. 4c, Supplementary Data 5) suggested involvement of AS genes in DNA-related processes (e.g., covalent chromatin modification or regulation of chromosome organization), while only one implied a muscle characteristic feature (i.e., regulation of cardiac muscle cell action potential). As we thought this observation might be biased due to the low number of GO terms detected in C2C12 (18), we repeated enrichment analysis, this time using a more relaxed threshold (non-corrected p-value < 0.01 instead of FDR < 0.01) on AS genes that would enter GO analysis. However, even among 45 enriched GO terms obtained using less stringent threshold, DNA-related processes comprised more than a third of all GO terms (36%, Supplementary Fig. 2h), which was not the case for NSC34 cells.
RBPs expressed in NSC34 and C2C12 influence TDP-43-dependent splicing
The observation that TDP-43 loss elicits a tissue-characteristic response did not come as a surprise, as RNA-binding proteins (RBPs) other than TDP-43 might be differentially expressed in these cells. Inspecting expression levels of some RNA-binding proteins14, which either directly interact with TDP-4340 or influence processing of its target transcripts16,31,41, we saw a higher average expression of RBPs in neuronal NSC34 cells (Fig. 5a, Supplementary Data 1) in line with previous observations14. Their joint functions in coordinating mRNA processing might underlie a more complex splicing regulation that is unique for neuronal tissues and explain why TDP-43-regulated splicing is more frequent in NSC34 than in C2C12 cells (Fig. 3). The two cell types clearly express a distinct array of RBPs (Fig. 5b, Supplementary Data 1), while abundance of some mRNAs is additionally affected by TDP-43 depletion (Fig. 5c, Supplementary Data 2).
a Boxplot shows that NSC34 cells on average display higher expression of 63 RNA-binding proteins14 compared to C2C12 cells (p-value = 0.0028). Average expression levels are plotted as log10-transformed FPKM values of all 63 transcripts and p-value was generated using Wilcoxon signed-rank test. b Expression of 63 RBPs (plotted as log10-transformed FPKM values) in C2C12 and NSC34 cells (Spearman’s ρ = 0.94; p-value < 2.2·10−16). Those with higher expression in one cell line than another (> 150%) are shown in red (C2C12) or blue (NSC34). Grey line represents y = x. c Venn diagram shows RBPs the expression of which changes following TDP-43 reduction. The overlapping event is downregulation of Tardbp.
Having identified and validated the most representative cell-type-specific AS events (Fig. 6b), we next explored whether TDP-43-dependent expression of specific splicing isoforms could be explained by the co-regulatory activity of other RBPs that are highly expressed in one cell line but are absent (or have negligible levels) in the other. Using siRNA against these proteins, we knocked down four RBPs that are most prominently expressed in a cell-type-specific manner (i.e., Elavl4 and Elavl3 in NSC34; Celf2 and Khdrbs3 in C2C12) (Fig. 6a). As a consequence, we could see that in three events out of the six tested, the expression of tissue-characteristic TDP-43-regulated splice isoforms (Fig. 6b, c) indeed depends on the co-regulation by these tissue-specific splicing actors. For example, by silencing muscle-characteristic RBPs Celf2 or Khdbrs3 in C2C12 cells, we decreased the expression of the long Tbc1d1 isoform typical of C2C12 cells and obtained a more “neuron-like” phenotype even in the presence of TDP-43 (Fig. 6d). By silencing Elavl3 or Elavl4 in NSC34 cells, on the other hand, we could shift the ratio of Dync1i2 and Lrp8 splice isoforms expressed in NSC34 cells towards the isoform typically present in C2C12 cells (Fig. 6e). Although a more comprehensive analysis of the role played by all tissues-specific splicing factors (also in combination with one another) would be necessary to obtain a complete picture, these results show that the cell-type-specific splicing activity of TDP-43 arises as consequence of different cellular environments, and especially the characteristic expression of tissue-specific RBPs that modulate inclusion of selected TDP-43-targeted exons.
a Expression levels of four RBPs following their knock-down in C2C12 and NSC34 cells (Celf2 and Khdrbs3 were silenced in C2C12; Elavl3 and Elavl4 were silenced in NSC34) as assessed using qPCR. p-values were generated using Student’s t-test (paired, two-tailed, n = 3 per group). b C2C12-specific and c NSC34-specific TDP-43-regulated splicing. Semi-quantitative RT-PCR conducted in TDP-43-silenced samples and corresponding controls is shown along with the quantification of splicing changes (% of alternative exon inclusion). The number of the alternative exon is shown in the scheme (see the exact transcript numbers in Supplementary Table 1). Coverage track from Interactive genome viewer (IGV) is shown on the right. d Splicing of C2C12-specific and e NSC34-specific TDP-43 targets depends on the presence of other tissue-specific factors. RT-PCR was conducted in samples silenced for specific RBPs (Celf2 and Khdrbs3 in C2C12; and Elavl3 and Elavl4 in NSC34 cells) together with corresponding controls. Quantification of splicing changes is shown below (% of alternative exon inclusion). Significance was tested using Student’s t-test (paired, two-tailed, n = 3 per group).
Common TDP-43 splicing targets detected in C2C12 and NSC34
Previous studies have already disclosed lists of transcripts, whose splicing is affected by TDP-43 removal or dysfunction21,24,28,41. Yet, the reproducibility of target identification is rather poor, possibly due to differences in methodological approaches, low conservation of TDP-43 targets across species28, and, as we show, the unique function TDP-43 elicits in each tissue or celltype. The most consistently reported TDP-43-regulated splicing event across studies and conditions is skipping of exon 3 within Poldip3/POLDIP3 mRNA (both mouse and human)23. This being so, inclusion level (percent spliced in, ΔPSI) of Poldip3 exon 3 often serves as a readout of TDP-43 functionality42,43,44. In search of new splicing events that would, similarly to Poldip3/POLDIP3, show high reproducibility across experimental settings, we chose mRNAs that underwent the biggest shift in TDP-43-dependent exon inclusion and whose isoform proportion was altered in both C2C12 and NSC34 cells. The isoform switch of these targets was validated using isoform-sensitive semiquantitative RT-PCR (Fig. 7a).
a Validation of TDP-43 dependent splicing of 10 representative mRNA targets. Semi-quantitative RT-PCR conducted in TDP-43-silenced samples and corresponding controls is shown along with the quantification of splicing changes (% of alternative exon inclusion). The number of the alternative exon is shown in the scheme (see the exact transcript numbers in Supplementary Table 1, n = 3 replicates per group). b Average expression levels of transcripts that are commonly spliced in both cell lines (164) or in one cell line exclusively (1268) is plotted as log10-transformed FPKM values (p-value < 2.2 · 10−16). c Absolute changes (∆PSI) of overlapping splicing events (100) compared to those uniquely occurring in C2C12 or NSC34 (1800) (p-value = 1.0 · 10−7). P-values for b, c were generated by unpaired Wilcoxon rank-sum test. d The correlation of splicing changes for commonly detected splicing events (100) plotted as ∆PSI in C2C12 and NSC34 (Spearman’s correlation coefficient ρ = 0.62, p-value = 4.1 · 10−12). e Percentage of frame-preserving AS events among those that commonly occur in both cell lines (100) and those regulated by TDP-43 in a cell-type specific manner (1800). f Average per exon PhyloP conservation scores plotted as box plots show TDP-43-regulated alternative sequences detected in both cell lines (634) are better conserved across species than those detected in one cell line exclusively (6019) (p-value = 0.02). P-value was generated using Wilcoxon rank-sum test, the grey line represents the median of average PhyloP scores of all exons in the mouse genome.
Relative to cell-type-specific TDP-43 targets, commonly spliced transcripts on average show higher expression in C2C12 and NSC34 cells than transcripts alternatively spliced in a cell-type-specific manner (Fig. 7b, Supplementary Data 1). Furthermore, commonly detected events display bigger splicing transitions (bigger ΔPSI) (Fig. 7c, Supplementary Data 3). Most of the splicing changes detected in C2C12 and NSC34 occurred in the same direction (83%, ρ = 0.62, p-value < 0.001) (Fig. 7d, Supplementary Data 3), meaning that for that subset of transcripts, TDP-43 exerts a similar function in cells of neuronal and muscular background. We observed a higher frequency of frame-preservation among splicing events found to be controlled by TDP-43 in both cell lines (Fig. 7e, Supplementary Data 3) along with better conservation of common TDP-43-regulated sequences across species (Fig. 7f).
We further compared results obtained in TDP-43-silenced cells (NSC34 and C2C12) to an RNA-seq dataset previously generated by Fratta et al.45, in which they sequenced brain tissue of mice carrying a mutation in important RNA-recognition motif RRM2 (F210I) of endogenous Tardbp gene leading to partial loss of TDP-43’s splicing function (Supplementary Fig. 3).
That dataset was subjected to the same MAJIQ pipeline for detection of alternatively used splicing junctions (Fig. 3c, Supplementary Data 4). The overlap between splicing events detected in TDP-43-silenced NSC34 cells and brain tissue of mice with the RRM2 mutation was 10.1% compared to 15.2% of splicing events commonly identified in C2C12 and NSC34 following TDP-43 knockdown.
This may seem rather low, however, several considerations must be made: first, cell lines (C2C12 or NSC34, respectively) consist of homogenous cell populations, which is in contrast to heterogeneous cell composition within bulk brain tissue. Second, different from embryonic mouse brain (E18.5) containing neuronal cells at different stages of differentiation, only undifferentiated cells were used in our study (C2C12 and NSC34). It should also not be neglected that TDP-43 depletion in C2C12 and NSC34 cells was achieved by siRNA transfection whilst there was some functional TDP-43 still present in the F210I embryonic brain (as complete knockout of TDP-43 in mouse embryo leads to a very early embryonic lethality before day 12 of embryogenesis)46.
We believe that results generated by a comparative analysis of TDP-43’s function in NSC34 and C2C12 cells, that were chosen as two cell lines representing muscle and neurons, clearly demonstrate that the activity of TDP-43 itself depends on the cellular context. Nonetheless, NSC34 cells are used in undifferentiated state and can only to a limited extent recapitulate (structural and functional) features of mouse brain. This being said, we believe that a 10.1% overlap consistently supports the idea that there is a small subset of transcripts targeted by TDP-43 across different cell types (like for example Poldip3, Ppfibp1 and Tmem2) while splicing of others strongly depends on cell-specific factors.
TDP-43-regulated splicing events conserved between mouse and human
While incorporation of exon 3 into mature Poldip3 mRNA is regulated by TDP-43 in both mouse and human cells23, most of TDP-43’s regulated splicing has been shown to be highly species- and even cell-type-specific. We therefore investigated if any of commonly detected TDP-43 targets (Fig. 7a) are (according to VastDB47) predicted to have an orthologous event in humans. Some TDP-43-mediated events found in mice (Rgp1 exon 3, Sapcd2 exon 2, Fam220a exon 2) do not even have a corresponding orthologous exon in humans. For those with putative AS orthology (i.e., the presence of orthologous alternative exon in both species), we tested whether alternative exons were subject to TDP-43 control also in human cells. We silenced TDP-43 in two human cell lines representing neuronal and muscular cells — human neuroblastoma SH-SY5Y and rhabdomyosarcoma RH-3048 (Fig. 8a), which resulted in exon skipping within POLDIP3 (Fig. 8b). Likewise, TDP-43 depletion led to enhanced inclusion of exon 19 in PPFIBP1 and exon 23 of ASAP2 but not exon 5 of TRAF7 or exon 3 of NFYA (Fig. 8c). Although one study reported a great portion of TDP-43-controlled exons in mouse to have a prior evidence of alternative splicing in humans20 we still lack understanding to what extent TDP-43 regulation of mRNA processing is conserved between species. Exon orthology (as assessed by sequence similarity) could not be predictive of AS conservation since exon incorporation into mature mRNA depends on the exonic sequence but also on cis-regulatory motives and trans-acting factors49,50,51. In fact, iCLIP performed in SH-SY5Y cells21 identified direct TDP-43-binding sites a in close proximity of alternatively spliced exons within PPFIBP1 and ASAP2, while that was not the case for TRAF7 and NFYA (Fig. 8d). This finding suggests that alternative exons of PPFIBP1 and ASAP2 found to be regulated by TDP-43 in mouse and human cells are most likely controlled by TDP-43 in a direct fashion by its binding to regulatory sequences neighbouring splice sites.
a Western blot shows efficient reduction of TDP-43 in SH-SY5Y and RH-30 cells upon siTDP transfection. The amount of TDP-43 was normalized against GAPDH or tubulin (n = 3 replicates per group). b TDP-43 depletion led to altered splicing of POLDIP3. Semi-quantitative RT-PCR conducted in TDP-43-silenced samples and corresponding controls is shown along with the quantification of splicing changes (% of alternative exon inclusion). The number of the alternative exon is given below (n = 3 replicates per group). c Alternatively spliced exons regulated by TDP-43 in mouse cells are either subject to TDP-43 regulation in human cells (PPFIBP1 exon 19 and ASAP2 exon 23) or not (TRAF7 exon 5 and NFYA exon 3). Semi-quantitative RT-PCRs conducted in TDP-43-silenced samples and corresponding controls are shown along with the quantification of splicing changes (% of alternative exon inclusion). Number of the alternative exon is given in the scheme (see the exact transcript numbers in Supplementary Table 2, n = 3 replicates per group). d Schematic representation of TDP-43 binding sites identified by iCLIP analysis in SH-SY5Y cells21 in the vicinity of exons represented on c.
Altered splicing patterns imply on TDP-43 dysfunction in FTLD and IBM patients
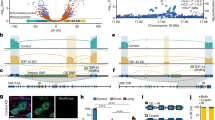
To explore if dysregulated alternative splicing could play a role in the pathophysiology of TDP-43 proteinopathies, we measured inclusion levels of TDP-43-mediated alternative exons in IBM muscles (Fig. 9a, Supplementary Data 6) as well as in pathological brain regions of ALS and FTLD cases with reported TDP-43 pathology (ALS-TDP and FTLD-TDP) (Fig. 9b, c, Supplementary Data 6). Since neuroanatomical regions markedly vary with regards to splice isoform expression (Supplementary Fig. 4a), we considered each brain region independently rather than analysing them together. Tissue-specific accumulation of truncated STMN2, which has recently been described as a very good clinical marker of TDP-43 impairment22,43,52, in fact occurs in brain areas previously known to be affected by TDP-43 pathology. We thus investigated TDP-43-controlled splicing in the spinal cord (lumbar and cervical, respectively) and the motor cortex of ALS cases (Fig. 9b, Supplementary Data 6), whereas frontal and temporal cortices were the site of interest for FTLD patients (Fig. 9c, Supplementary Data 6).
a Inclusion levels (PSI) of six alternative exons in skeletal muscle biopsies in IBM patients vs healthy controls (n = 4 per group). b PSI of six alternative exons in different brain regions (motor cortex, lumbar spinal cord, cervical spinal cord) of ALS patients and healthy controls (n motor cortex: 223 ALS and 23 ctrl, n cervical spinal cord: 134 ALS and 32 ctrl, n lumbar spinal cord 136 ALS and 33 ctrl). c PSI of six alternative exons in frontal and temporal cortices of FTLD patients with reported TDP-43 pathology and healthy controls (n frontal cortex: 33 FTLD and 40 ctrl, n temporal cortex: 30 FTLD and 23 ctrl). a–c P-values were generated using Wilcoxon rank-sum test. *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001. d Schematic summary of all splicing alterations a–c detected in skeletal muscles of IBM patients and across neuroanatomical regions of ALS and FTLD patients compared to healthy controls. Dark green marks significant changes, which occur in the same direction as in TDP-43-depleted SH-SY5Y and RH-30 cells (refers to Fig. 6c); light green marks non-significant changes that occur in the expected direction (p-value is reported in the scheme); red marks significant changes occurring in the opposite direction relative to TDP-43-depleted SH-SY5Y and RH-30 cells; light red marks non-significant changes that occur in the opposite direction (p-value is reported in the scheme).
Splicing signature examined herein consisted of six TDP-43-regulated alternative exons: POLDIP3 exon 3 is consistently detected as a TDP-43-regulated splicing event; exon 15 of TNIK has been previously described as TDP-43 target and was also detected in C2C12 (Fig. 6b), SH-SY5Y and RH-30 cells (Supplementary Fig. 4b); exon 19 of PPFIBP1 and exon 23 of ASAP2 are newly identified TDP-43 targets conserved across species; exons 12 and 13 of TBC1D1 are detected to be controlled by TDP-43 in C2C12 cells and are associated with muscle differentiation. The long TBC1D1 isoform, which is dependent on TDP-43, appears to be crucial in mature tissues but not in undifferentiated cells (hence, it was not detected in undifferentiated NSC34 (Fig. 6b), SH-SY5Y and RH-30 cells (Supplementary Fig. 4b)). As TDP-43 binding sites were indeed found in the proximity of these alterative exons (Supplementary Fig. 4c), inclusion levels of exons 12 and 13 of TBC1D1 gene were investigated in mature tissues coming from patients.
Interestingly, when we assessed inclusion levels of six TDP-43-controlled AS events in patient tissues, we got distinct patterns. For example, out of the six AS events, we only observed increased ASAP2 exon 23 inclusion in IBM muscle relative to healthy controls (Fig. 9a, Supplementary Data 6). In ALS cases, we detected significantly different inclusion of one exon (exon 19 of PPFIBP1) in the lumbar and cervical spinal cord but not in the motor cortex, while in motor cortex, we saw enhanced skipping of both alternative exons within TBC1D1 (Fig. 9b, Supplementary Data 6). Intriguingly, FTLD appears to be the disease in which splicing of six alternative exons is most heavily perturbed. Multiple TDP-43-targeted exons show significantly altered inclusion in patients, both in frontal and temporal cortices (Fig. 9c, Supplementary Data 6). Apart from that, some non-significant changes clearly show a trend towards altered exon inclusion in patients.
At this point it is important to consider cell-type-specific splicing activity of TDP-43 (Fig. 3a, Supplementary Data 3), which makes it unlikely that upon TDP-43 malfunction, splicing of the same transcripts would be altered across all cell-types (Fig. 9d). This being said, the scheme in Fig. 9d summarizes splicing changes of six TDP-43-controlled exons detected in different tissues affected with TDP-43 pathology. The fact that splicing changes do not necessarily occur in the same direction following TDP-43 depletion in cell lines (as in the case of ASAP2 and TNIK) again highlights the complexity of splicing control provided by TDP-43 that is generally acting within an interwoven network of splicing regulators. The same phenomenon (i.e., different directionality) was in fact observed when comparing the consequences TDP-43 depletion has on gene expression and alternative splicing in vitro using cell lines (Figs. 2c and 7d, Supplementary Data 2 and 3).
Discussion
TDP-43 inclusions represent the hallmark of ALS/FTLD5,7 and are frequently recognized as a secondary pathology in other neurodegenerative disease53,54. In recent years, great progress has been made in explaining how potential loss- and gain-of-function mechanisms contribute to the pathogenesis observed in the brain and spinal cord27,45,55. Nonetheless, a growing evidence of TDP-43 mis-localization and aggregation in tissues beyond the CNS has raised the possibility that TDP-43 dysfunction and consequently, impairment of RNA processing, might be deleterious for other tissues42,56.
To this date, cell- and tissue-characteristic molecular features of TDP-43 have seldom been investigated in parallel. Considering recent attention that TDP-43 has received in IBM and related pathologies8,9,11,57, we, therefore, sought to fill this gap. The purpose of our study has been to further explicate the role of TDP-43 in different tissues to better understand its involvement in pathogenesis in cell-types other than neurons, and to set the ground for development of potential therapeutic or biomarker strategies that focus on shared or specific disease mechanisms. We thus aimed to model protein loss/dysfunction in skeletal muscles vs. neurons and to focus on TDP-43-controlled alternative splicing (AS) events, as this is one of the best characterized features of this protein to date.
Although protein levels of TDP-43 itself are not different between C2C12 and NSC34, there is a tissue-characteristic expression of other RNA-binding proteins (e.g., those from Elavl, Nova and Celf families) that, like TDP-43, mediate RNA-related processes in a coordinated fashion. This result, together with differences in tissue-specific gene expression levels, can explain why there is little consistency across studies in identifying TDP-43-targeted transcripts58, and it clearly outlines the importance of cellular context in shaping the functional role of TDP-43. With regards to future TDP-43 investigations, our findings highlight the need to employ tissue models, which are most relevant for a certain condition. Most importantly, our results show that in the case of TDP-43 proteinopathies, the knowledge acquired by studying neuronal cells could be translated to muscles only to a limited extent. Despite not being investigated in this work, the same presumably applies for the interpretation of iCLIP results, in which TDP-43 binding should be always considered in the context of tissue characteristic environment, having in mind possible differences in binding behaviour of the protein across cell-types that might affect splicing59 and expression changes43.
This initial comparison of TDP-43’s function across celltypes based on RNA-seq discusses changes that occur at the mRNA level. With regards to mRNA splicing, it remains unclear whether TDP-43-dependent changes in mRNA isoforms actually translate in corresponding protein isoforms. This would require an extensive screening using isoform-specific antibodies or a mass-spectrometry-based approach. Nonetheless, as a proof-of-principle, we show that for TBC1D1, TDP-43-dependent ratio of two protein isoforms actually reflects that of Tbc1d1 mRNAs (Supplementary Fig. 5).
In some cases, our parallel study has given expected results. In NSC34 cells, for example, TDP-43 loss impacts expression of genes participating in pathways that provide elemental functions of neuronal cells, like vesicle-mediated transport and regulation of postsynaptic membrane neurotransmitters, which is perfectly in line with previous studies20,21. Similarly, the loss of TDP-43 in C2C12 cells impairs muscle-characteristic features, like striated muscle development, muscle cell migration or regulation of muscle cell differentiation, which has been functionally confirmed by others12,29. However, we have also detected cell-type-specific TDP-43-associated dysregulation of molecular functions that will probably deserve further investigation. For example, we found that in muscles TDP-43 mediates splicing of mRNAs encoding proteins implicated in DNA-related processes. This is a particularly interesting observation as DNA-related processes play an important role in muscle differentiation. In adult skeletal muscle, DNA and histone modifications participate in adaptive response to environmental stimuli, which challenge structural and metabolic demands and thus make skeletal muscle a very plastic tissue60,61. Also, the early commitment towards myogenic lineage involves epigenetic changes mediated by chromatin remodelling enzymes like histone deacetylases (HDACs), histone acetyltransferases (HATs) and histone methyltransferases (HMTs)62. In keeping with this, Dnmt3a, Dnmt3b, Hdac9, Hdac7, Prdm2 are just few of chromatin-modifying enzymes that underwent splicing changes upon TDP-43 depletion in C2C12 but not in NSC34 cells. Interestingly, telomere shortening was described in primary muscle cultures of sIBM patients suggesting premature senescence63 and epigenetic changes have been described in congenital myopathies64. Therefore, the results obtained in C2C12 suggest another possible mechanism on how TDP-43 may control gene expression in muscle in an indirect fashion and eventually participate in disease. Recently, loss of TDP-43 was associated with increased genomic instability and R-loop formation65,66, possibly through mechanisms involving Poldip3, which has been shown to play a role in maintaining genome stability and preventing R-loop accumulation at sites of active replication67.
On the other hand, some molecular processes such as dysregulation of protein assembly and disposal, mitochondrial changes and apoptosis; as well as alterations in post-translational modifications seem to occur upon TDP-43 depletion in both cell types, which possibly links these pathological changes to TDP-43 dysfunction in both tissues. As we have drawn our conclusions based on the RNA-seq analysis, a crucial future step will be to functionally assess to what extent TDP-43 loss impacts the above-mentioned processes in each tissue. Ideally, functional experiments should be performed in the two cell types in parallel, as only such approach would allow a direct comparison of the regulatory role played by TDP-43 in each context and would answer the question whether impairment of RNA processing is as central in IBM as it is in ALS.
Working with cell lines representing muscles12,29,48 and neurons21,23,28,59,68 allowed us a direct (and unbiased) assessment of TDP-43 activity across cel- types. Mouse cell lines have been routinely employed to study TDP-4312,29. In our case, they were chosen over human cells due to the lack of an appropriate and well-established cell line derived from human skeletal muscle. With regards to the contribution of TDP-43 malfunction to human pathology, we observed that transcripts, whose splicing was commonly affected by TDP-43 loss in the two mouse cell lines, appear more likely to undergo TDP-43-regulated processing also in human cells. Herein, we show that alternative sequences regulated by TDP-43 in both cell lines are better conserved between species than those regulated in a cell type-specific manner. Nonetheless, a conservation of the alternative sequence itself cannot guarantee for splicing conservation. Thus, it would be extremely insightful to investigate conservation of TDP-43-regulated splicing between humans and mice on a transcriptome-wide level by actual sequencing experiment (rather than comparing gene sequences as such), possibly using analogous tissues69. A good example of commonly regulated event is skipping of exon 3 within POLDIP3, the regulation of which is conserved between mouse in humans20,23,32 and has made it the most consistently detected event across studies. In this study, however, we identified two additional targets, ASAP2 and PPFIBP1, and show that they also indeed undergo TDP-43-dependent splicing in all (mouse and human) cell lines tested. These additional findings could be of interest to identify common endpoints of mouse and human disease models that could then be used to monitor the efficiency of eventual therapeutic approaches or to follow disease course/onset.
Finally, as a proof-of-principle, we show that splicing alterations of TDP-43-dependent transcripts do in fact take place in different tissues (i.e., skeletal muscle and certain brain regions) affected by TDP-43 pathology. While expression levels of a given transcript heavily vary between individuals and, in our experience, seem to be influenced by experimental procedure itself (how and when biopsies are taken), the relative abundance of characteristic isoforms appears to be a more reliable readout. Considering cell-type-specific activity of TDP-43, it is reasonable to deduce that splicing of other TDP-43-controlled transcripts would be affected in the skeletal muscle and in neurons. In conclusion, we show that splicing changes represent a robust indication of pathological conditions both in the skeletal muscle of IBM patients and in the brain of individuals affected with FTLD.
Methods
Cell culture
C2C12 immortalized mouse myoblasts (ECACC), SH-SY5Y human neuroblastoma (ECACC) and RH-30 human rhabdomyosarcoma (kindly donated by Marc-David Ruepp) were maintained in DMEM (Thermo Fisher Scientific), supplemented with 10% FBS (Thermo Fisher Scientific) and antibiotics/antimycotics (Sigma-Aldrich) under standard conditions. NSC34 motoneuron-like mouse hybrid cell line (available in house) was cultured in DMEM (Thermo Fisher Scientific) with 5% FBS (Sigma-Aldrich) and antibiotics/antimycotics (Sigma-Aldrich). All experiments were performed with cells of similar passage number (± 2). To silence RNA-binding proteins (TDP-43, Celf2, Khdrbs3, Elavl3, Elavl4) in C2C12 and NSC34 cells, 40 nM of siRNA (mouse siTDP 5'-CGAUGAACCCAUUGAAAUA-3', Sigma-Aldrich, pre-designed mouse siCelf2, siKhdrbs3, siElavl3 and Elavl4, Thermo Fisher Scientific) or non-targeting siLUC (5'-UAAGGCUAUGAAGAGAUAC-3', Sigma-Aldrich) were mixed with 54 μl of RNAiMAX (Invitrogen) following manufacturer’s reverse transfection protocol and applied to 700,000 cells seeded in a 10 cm dish. 48 h later, transfected cells were collected for subsequent analysis. The same reagent was used to silence TDP-43 in human SH-SY5Y and RH-30 cells. 400,000 RH-30 were seeded in a 6 cm dish, reversely transfected (human siTDP 5’-GCAAAGCCAAGAUGAGCCU-3’, Sigma-Aldrich or siLUC) and harvested 48 h later. To deplete TDP-43 in SH-SY5Y cells, 10,00,000 cells were seeded in a 6 cm dish and reversely transfected. After 48 h, they were transfected again and harvested 48 h later.
Western blotting
Whole-cell extracts were resuspended in PBS in the presence of protease inhibitor and sonicated. 15 μg of protein sample were separated on a 10% Bis-Tris gel (Invitrogen) and transferred to a nitrocellulose membrane (Invitrogen). The membrane was blocked in 4% milk-PBST (PBS with 0.1% Tween-20) and proteins were stained using the following antibodies: anti-TDP-43 (rabbit, Proteintech, 1:1000), anti-GAPDH (rabbit, Proteintech, 1:1000), anti-HSP70 (rat, EnzoLife Science, 1:1000), anti-tubulin (mouse, available in house, 1:10,000) and HRP-conjugated secondary antibodies anti-rabbit (goat, Dako, 1:2000), anti-mouse (goat, Dako, 1:2000), anti-rat (rabbit, Dako, 1:2000).
To detect TBC1D1, 30 μg of protein sample were separated using a pre-cast gradient gel (4–12%, Invitrogen) and wet blotted onto a PVDF membrane (Merck Millipore). The membrane was then blocked in 4% milk-TBST, stained with anti-TBC1D1 (rabbit, Cell Signalling, 1:1 000, in 2% milk-TBST) and anti-rabbit HRP-conjugated secondary antibody (goat, Dako, 1:2 000).
Image acquisition and result quantification were conducted using Alliance Q9 Advanced Chemiluminescence Imager (UviTech).
RNA extraction, RT-PCR
Total RNA was isolated using standard phenol-chloroform extraction. RNA quality was assessed using capillary electrophoresis (Qiaxcel RNA QC Kit v2.0, Qiaxcel) and quantified using Qiaxcel software (QIAxcel ScreenGel (v1.4.0)). Only high-quality (RIS > 8) RNA of high purity (A260/A230 and A260/A280 > 1.8) was taken for subsequent analysis. 500 ng of RNA were reversely transcribed using random primers (Eurofins) and Moloney murine leukaemia virus reverse transcriptase (M-MLV, Invitrogen) according to manufacturer’s instructions.
Splicing-sensitive PCR and qPCR
For detection of alternatively spliced mRNAs, PCR primers were designed complementary to constitutive exonic regions flanking a predicted alternatively spliced cassette exon. PCR mix was prepared using gene-specific primers (0.6 μM, Sigma, primer sequences in Supplementary Tables 1 and 2) and TAQ DNA polymerase (Biolabs or Roche) according to manufacturer’s instructions and subjected to PCR protocol (35–45 amplification cycles) optimized for each primer pair. PCR products were separated by capillary electrophoresis (DNA screening cartridge, Qiaxcel), and splicing transitions were quantified using Qiaxcel software (Qiaxcel ScreenGel (v1.4.0)). Exon inclusion was calculated by the software. Percentage of the inclusion (Inc. %) reports the area under the curve of the peak representing the longer (inclusion) splicing isoform.
For assessment of transcript levels, real-time quantitative PCR was performed using PowerUp SYBR Green master mix (Applied Biosystems) and gene-specific primers (primer sequences in Supplementary Table 3). cDNA was subjected to 45 cycles of the following thermal protocol: 95 °C for 3 min, 95 °C for 10 s, 65 °C for 30 s, 95 °C for 10 s, 65 °C for 1 s. Relative gene expression levels were determined using QuantStudio design and analysis software (Thermo Fisher Scientific (v1.5.1)) always comparing treated samples (siTDP) with their direct controls (siLUC) normalized against Gapdh. P-values were calculated using one-tailed paired t-test as qPCR was conducted to validate expression changes detected by RNA-seq.
RNA-seq
Both polyA cDNA library generation and RNA-seq were performed by Novogene (Beijing, China). cDNA libraries with insert lengths of 250–300 bp were generated using NEB NextUltra RNA Library Prep Kit. Sequencing was conducted on Illumina HiSeq 2500 with paired-end 150 bp (PE 150) strategy.
Read mapping
Sequencing quality control and filtering were performed to prune reads with average Phred score (Q score) below 20 across 50% of bases, as well as those with > 0.1% of undetermined (N) ones. Obtained reads were aligned to the mouse genome GRCm38 (mm10) using the Spliced Transcripts Alignment to a Reference (STAR) software (v2.5)70, an RNA-seq data aligner that utilizes Maximal Mappable Prefix (MMP) strategy to account for the exon junction problem.
Quantification of gene expression level
Counting of reads mapped to each gene was performed using HTSeq (v0.6.1)71. Raw read counts together with respective gene length were used to calculate Fragments Per Kilobase of transcript sequence per Million base pairs sequenced (FPKM). In contrast to read counts, FPKMs account for sequencing depth and gene length on counting of fragments72 and are frequently used to estimate gene expression levels.
Differential expression analysis
Differential gene expression (DEG) analysis of two conditions was performed using the DESeq2 R package (v2.1.6.3)73, a tool that utilizes negative binomial distribution model to account for variance-mean dependence in count data and tests for differential expression74. Three biological replicates were included per cell-type and condition, in control (siLUC) and TDP-43-silenced (siTDP) cells. Read count matrix was pre-filtered by removing rows with row sum below 1. Multiple testing adjustments were performed using Benjamini and Hochberg’s approach to control for the false discovery rate (FDR). Transcripts with padj < 0.05 were considered as differentially expressed.
Differentially expressed genes identified in both cell lines under different experimental conditions were hierarchically clustered based on log10(FPKM + 1) and visualized with pheatmap R package (v1.0.12)75. Further, distance between silenced and control samples of each cell line was illustrated with principal component analysis (PCA), using the R function “prcomp”76. Differences in gene expression levels (log10(FPKM + 1)) between cell lines were tested for significance using Wilcoxon signed-rank test.
Alternative splicing analysis
Five major types of alternative splicing events — skipped exons (SE), mutually exclusive exons (MXE), alternative 5' and 3' splice sites (A5'SS and A3'SS) and intron retention (RI) — were detected and analysed by Novogene using replicate multivariate analysis of transcript splicing (rMATS) software (v3.2.1)77. Every alternative splicing event can produce exactly two isoforms. Each isoform is adjusted for its effective length before calculating the ratio of two isoforms and testing significance of differential splicing between two conditions. Multiple testing was corrected using Benjamini and Hochberg’s method. Splicing events having FDR < 0.01 were considered significant irrespective of ΔPSI.
Alternatively (for analysis of cryptic splicing and patient’s data (Figs. 3c and 9, Supplementary Data 4, Supplementary Figs. 3 and 4a)), differential splicing analysis was performed using MAJIQ (v2.1) and the GRCm38 as a reference genome as previously described elsewhere78.
Enrichment analysis
Gene Ontology GO79 is widely used in gene enrichment analysis to classify list of individual genes based on their expression pattern, or other similar feature, with the aim to predict dysregulated biological processes, functions and pathways or any other general trend within a subset of data80. In this study, GO enrichment analysis was conducted in R, using clusterProfiler package (v3.14.3)80 either on the set of differentially expressed genes (padj < 0.05) or alternatively spliced genes (FDR < 0.01), if not stated otherwise. Additionally, GO enrichment analysis was conducted using a less stringent threshold for inclusion of alternatively spliced genes (where we considered genes with non-corrected p-value < 0.01 instead of FDR < 0.01). Genes of a particular dataset were assigned Entrez gene identifiers from Bioconductor mouse annotation package org.Mm.eg.db (v3.10.0). Enrichment test for GO terms was calculated based on the hypergeometric distribution. The resulting GO terms/KEGG pathways were considered significant after applying multiple testing corrections with Benjamini–Hochberg method (padj < 0.05). Subsequently, significant GO terms (category: biological process) were functionally grouped or manually edited depending on the underlying biological question.
Conservation analysis
Gene/exon conservation analysis within mouse (mm10) was performed by calculating phyloP (phylogenetic p-values) scores, i.e., per base conservation scores, generated from aligned genomic sequences of multiple species81.
For each differentially expressed gene, the average per gene phyloP score was computed with bigWigSummary (UCSC)82. To calculate phyloP scores of TDP-43-regulated alternative sequences (hereafter referred to as per exon phyloP score), we considered TDP-43-regulated sequences of all event types. Those include A’3SS and A’5SS (long and short exon), retained introns and cassette exons (SE, the 1st and the 2nd exon of MXE).
Patient samples
The NYGC ALS cohort has previously been detailed elsewhere22,78. Herein, we only considered ALS and FTLD patients with TDP-43 pathology (ALS-TDP and FTLD-TDP) and healthy controls while excluding ALS with SOD1 mutations of FTLD patients without TDP-43 inclusions.
The NYGC ALS Consortium samples presented in this work were acquired through various Institution Review Board protocols from member sites and the Target ALS post-mortem tissue core. They were transferred to the NYGC in accordance with all applicable foreign, domestic, federal, state, and local laws and regulations for processing, sequencing, and analyses.
Muscle biopsies (vastus lateralis or biceps) were obtained from four patients diagnosed with IBM according to the Griggs criteria83 and four healthy controls. Participants were investigated for cramps or fatigue; they underwent regular examination, neurophysiology tests and histological examinations. IBM biopsies were taken from moderately affected muscles and routinely investigated for histological and immunohistochemistry features. In case muscle fibrosis was present, it did not compromise a definite pathologic diagnosis. Basic demographic features of all participants are summarised in Supplementary Table 4. Biopsies were stored at −80 °C. Institutional board reviewed the study and ethical approval was obtained.
Sample processing, library preparation, and RNA-seq quality control have already been described elsewhere78.
Statistics and reproducibility
Statistical analysis was performed using R (v3.6.1) or a specific R package as described in dedicated sections above (see Quantification of gene expression level, Differential expression analysis, Alternative splicing analysis, Enrichment analysis and Patient samples). A significance level of 0.05 was generally considered for the analysis unless otherwise stated. The exact sample size and statistical tests used to test significance of the differerence are given in captions, along with the description of corrections and statistical parameters that are shown in each figure.
At least three biological replicates (experiments conducted at different time points) with technical triplicates were used for qPCR and western blot quantification.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Datasets generated for this study were deposited in NCBI’s GEO and are accessible through GEO Series accession number GSE171714. iCLIP data generated by Tollervey et al.21 that support the findings of this study were deposited in the ArrayExpress archive and are accessible at E-MTAB-530. RNA-seq data of the F210I mouse generated by ref. 45 were deposited in the NCBI Sequence Read Archive and are accessible at SRP133158. RNA-seq data generated through the NYGC ALS Consortium used in this study can be accessed at GEO (GSE137810, GSE124439, GSE116622 and GSE153960). To request immediate access to new and ongoing data generated by the NYGC ALS Consortium for samples provided through the Target ALS post-mortem core, contact ALSData@nygenome.org. All RNA-seq data in the NYGC ALS Consortium are made immediately available to all members of the Consortium and with other Consortia with whom we have a reciprocal sharing arrangement. Uncropped and unedited blot images are included in Supplementary Fig. 6. All data generated for this study is available from the corresponding author on reasonable request.
References
Birsa, N., Bentham, M. P. & Fratta, P. Cytoplasmic functions of TDP-43 and FUS and their role in ALS. Semin. Cell Dev. Biol. 99, 193–201 (2020).
Budini, M. & Buratti, E. TDP-43 autoregulation: implications for disease. J. Mol. Neurosci. 45, 473–479 (2011).
Ederle, H. & Dormann, D. TDP-43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett. 591, 1489–1507 (2017).
Buratti, E. & Baralle, F. E. TDP-43: gumming up neurons through protein–protein and protein–RNA interactions. Trends Biochem. Sci. 37, 237–247 (2012).
Arai, T. et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 (2006).
Buratti, E. Functional significance of TDP-43 mutations in disease. Adv. Genet. 91, 1–53 (Elsevier, 2015).
Neumann, M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006).
Salajegheh, M. et al. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis: redistribution of TDP-43. Muscle Nerve 40, 19–31 (2009).
Weihl, C. C. et al. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J. Neurol., Neurosurg. Psychiatry 79, 1186–1189 (2008).
Yamashita, S. et al. Optineurin is potentially associated with TDP-43 and involved in the pathogenesis of inclusion body myositis: Optineurin in inclusion body myositis. Neuropathol. Appl. Neurobiol. 39, 406–416 (2013).
Harms, M. B. et al. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann. Neurol. 71, 407–416 (2012).
Vogler, T. O. et al. TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature 563, 508–513 (2018).
Buratti, E. TDP-43 post-translational modifications in health and disease. Expert Opin. Ther. Targets 22, 279–293 (2018).
Mele, M. et al. The human transcriptome across tissues and individuals. Science 348, 660–665 (2015).
Appocher, C. et al. Major hnRNP proteins act as general TDP-43 functional modifiers both in Drosophila and human neuronal cells. Nucleic Acids Res. 45, 8026–8045 (2017).
Cappelli, S., Romano, M. & Buratti, E. Systematic analysis of gene expression profiles controlled by hnRNP Q and hnRNP R, two closely related human RNA binding proteins implicated in mRNA processing mechanisms. Front. Mol. Biosci. 5, 1–17 (2018).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011).
Jiang, C. et al. Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget 7, 7120–7133 (2016).
Ludwig, N. et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 44, 3865–3877 (2016).
Polymenidou, M. et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 (2011).
Tollervey, J. R. et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458 (2011).
Prudencio, M. et al. Truncated stathmin-2 is a marker of TDP-43 pathology in frontotemporal dementia. J. Clin. Investig. 130, 6080–6092 (2020).
Fiesel, F. C., Weber, S. S., Supper, J., Zell, A. & Kahle, P. J. TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 40, 2668–2682 (2012).
De Conti, L. et al. TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic and mitotic cellular pathways. Nucleic Acids Res 43, 8990–9005 (2015).
Prpar Mihevc, S., Baralle, M., Buratti, E. & Rogelj, B. TDP-43 aggregation mirrors TDP-43 knockdown, affecting the expression levels of a common set of proteins. Sci. Rep. 6, 1–9 (2016).
Prudencio, M. et al. Misregulation of human sortilin splicing leads to the generation of a nonfunctional progranulin receptor. Proc. Natl Acad. Sci. USA 109, 21510–21515 (2012).
Budini, M., Romano, V., Quadri, Z., Buratti, E. & Baralle, F. E. TDP-43 loss of cellular function through aggregation requires additional structural determinants beyond its C-terminal Q/N prion-like domain. Hum. Mol. Genet. 24, 9–20 (2015).
Colombrita, C. et al. TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochemistry 111, 1051–1061 (2009).
Militello, G. et al. A novel long non-coding RNA Myolinc regulates myogenesis through TDP-43 and Filip1. J. Mol. Cell Biol. 10, 102–117 (2018).
Jeong, Y. H. et al. Tdp-43 cryptic exons are highly variable between cell types. Mol. Neurodegener. 12, 1–9 (2017).
Mohagheghi, F. et al. TDP-43 functions within a network of hnRNP proteins to inhibit the production of a truncated human SORT1 receptor. Hum. Mol. Genet. 25, 534–545 (2016).
Shiga, A. et al. Alteration of POLDIP3 splicing associated with loss of function of TDP-43 in tissues affected with ALS. PLoS One 7, 1–11 (2012).
Askanas, V., Engel, W. K. & Nogalska, A. Sporadic inclusion-body myositis: a degenerative muscle disease associated with aging, impaired muscle protein homeostasis and abnormal mitophagy. Biochim. Biophys. Acta 1852, 633–643 (2015).
Askanas, V., Engel, W. K. & Nogalska, A. Pathogenic considerations in sporadic inclusion-body myositis, a degenerative muscle disease associated with aging and abnormalities of myoproteostasis. J. Neuropathol. Exp. Neurol. 71, 680–693 (2012).
Arnold, E. S. et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl Acad. Sci. 110, 736–745 (2013).
Irimia, M. et al. A highly conserved program of neuronal microexons Is misregulated in autistic brains. Cell 159, 1511–1523 (2014).
Yeo, G., Holste, D., Kreiman, G. & Burge, C. B. Variation in alternative splicing across human tissues. Genome Biol. 102, 2850–2855 (2004).
Green, C. J., Gazzara, M. R. & Barash, Y. MAJIQ-SPEL: web-tool to interrogate classical and complex splicing variations from RNA-Seq data. Bioinformatics 34, 300–302 (2018).
Mehmood, A. et al. Systematic evaluation of differential splicing tools for RNA-seq studies. Brief. Bioinforma. 21, 2052–2065 (2020).
Freibaum, B. D., Chitta, R. K., High, A. A. & Taylor, J. P. Global analysis of TDP-43 interacting proteins eeveals strong association with RNA splicing and translation machinery. J. Proteome Res. 9, 1104–1120 (2010).
Lagier-Tourenne, C. et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 15, 1488–1497 (2012).
Cortese, A. et al. Altered TDP-43-dependent splicing in HSPB8-related distal hereditary motor neuropathy and myofibrillar myopathy. Eur. J. Neurol. 25, 154–163 (2018).
Klim, J. R. et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat. Neurosci. 22, 167–179 (2019).
Roczniak-Ferguson, A. & Ferguson, S. M. Pleiotropic requirements for human TDP-43 in the regulation of cell and organelle homeostasis. Life Sci. Alliance 2, 1–12 (2020).
Fratta, P. et al. Mice with endogenous TDP‐43 mutations exhibit gain of splicing function and characteristics of amyotrophic lateral sclerosis. EMBO J. 37, 1–15 (2018).
Kraemer, B. C. et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 119, 409–419 (2010).
Tapial, J. et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 27, 1759–1768 (2017).
Reber, S. et al. Minor intron splicing is regulated by FUS and affected by ALS-associated FUS mutants. EMBO J. 35, 1504–1521 (2016).
Barbosa-Morais, N. L. et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 338, 1587–1593 (2012).
Gueroussov, S. et al. An alternative splicing event amplifies evolutionary differences between vertebrates. Science 349, 868–873 (2015).
Raj, B. & Blencowe, B. J. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron 87, 14–27 (2015).
Melamed, Z. et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 22, 180–190 (2019).
Hasegawa, M. et al. TDP-43 is deposited in the Guam parkinsonism-dementia complex brains. Brain 130, 1386–1394 (2007).
Higashi, S. et al. Concurrence of TDP-43, tau and α-synuclein pathology in brains of Alzheimer’s disease and dementia with Lewy bodies. Brain Res. 1184, 284–294 (2007).
Cascella, R. et al. Quantification of the relative contributions of loss-of-function and gain-of-function mechanisms in TAR DNA-binding protein 43 (TDP-43) proteinopathies. J. Biol. Chem. 291, 19437–19448 (2016).
Cortese, A. et al. Widespread RNA metabolism impairment in sporadic inclusion body myositis TDP43-proteinopathy. Neurobiol. Aging 35, 1491–1498 (2014).
Yamashita, S. et al. CYLD dysregulation in pathogenesis of sporadic inclusion body myositis. Sci. Rep. 9, 1–12 (2019).
Buratti, E., Romano, M. & Baralle, F. E. TDP-43 high throughput screening analyses in neurodegeneration: Advantages and pitfalls. Mol. Cell. Neurosci. 56, 465–474 (2013).
Highley, J. R. et al. Loss of nuclear TDP-43 in amyotrophic lateral sclerosis (ALS) causes altered expression of splicing machinery and widespread dysregulation of RNA splicing in motor neurones: Amyotrophic lateral sclerosis, TDP-43 and mRNA splicing. Neuropathol. Appl. Neurobiol. 40, 670–685 (2014).
Barrès, R. et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 15, 405–411 (2012).
McGee, S. L. & Hargreaves, M. Histone modifications and exercise adaptations. J. Appl. Physiol. 110, 258–263 (2011).
Guasconi, V. & Puri, P. L. Chromatin: the interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 19, 286–294 (2009).
Morosetti, R. et al. Increased aging in primary muscle cultures of sporadic inclusion-body myositis. Neurobiol. Aging 31, 1205–1214 (2010).
Rokach, O. et al. Epigenetic changes as a common trigger of muscle weakness in congenital myopathies. Hum. Mol. Genet. 24, 4636–4647 (2015).
Giannini, M. et al. TDP-43 mutations link amyotrophic lateral sclerosis with R-loop homeostasis and R loop-mediated DNA damage. PLoS Genet 16, 1–23 (2020).
Wood, M. et al. TDP-43 dysfunction results in R-loop accumulation and DNA replication defects. J. Cell Sci. 133, 1–14 (2020).
Björkman, A. et al. Human RTEL1 associates with Poldip3 to facilitate responses to replication stress and R-loop resolution. Genes Dev. 34, 1065–1074 (2020).
Nonaka, T. et al. Phosphorylated and ubiquitinated TDP-43 pathological inclusions in ALS and FTLD-U are recapitulated in SH-SY5Y cells. FEBS Lett. 583, 394–400 (2009).
Cardoso-Moreira, M. et al. Developmental gene expression differences between humans and mammalian models. Cell Rep. 33, 1–13 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, 1–12 (2010).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Kolde, R. pheatmap: Pretty Heatmaps https://cran.r-project.org/web/packages/pheatmap/index.html (2019).
R Core Team. R: A Language and Environment for Statistical Computing https://www.r-project.org/ (2019).
Shen, S. et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl Acad. Sci. USA 111, 5593–5601 (2014).
Brown, A.-L. et al. Common ALS/FTD Risk Variants in UNC13A Exacerbate its Cryptic Splicing and Loss Upon TDP-43 Mislocalization https://doi.org/10.1101/2021.04.02.438170 (2021).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29 (2000).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: An R Package for comparing biological themes among gene clusters. OMICS: A J. Integr. Biol. 16, 284–287 (2012).
Pollard, K. S., Hubisz, M. J., Rosenbloom, K. R. & Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010).
Kent, W. J., Zweig, A. S., Barber, G., Hinrichs, A. S. & Karolchik, D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207 (2010).
Griggs, R. C. et al. Inclusion body myositis and myopathies. Ann. Neurol. 38, 705–713 (1995).
Acknowledgements
We thank Marc-David Ruepp (King’s College London) for providing RH-30 cells and Robert Bakarić for his kind assistance with the conservation analysis. We would also like to thank the Target ALS Human Postmortem Tissue Core (New York Genome Center for Genomics of Neurodegenerative Disease, Amyotrophic Lateral Sclerosis Association) for providing post-mortem brain samples, patients and their families who donated those samples. IBM muscle biopsies were kindly provided by the Bank of muscle tissue, peripheral nerve, DNA and Cell Culture, a member of Telethon Network of Genetic biobanks, at Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico, Milano, Italy and from the Laboratory of Muscle Histopathology and Molecular Biology at IRCCS Policlinico San Donato, San Donato Milanese, Italy. This research was supported by the AriSLA grant PathensTDP to E. Bur. and by the European Reference Network for Neuromuscular Diseases to M. M. and M. Ro. Consortium activities were supported by the ALS Association (15-LGCA-234) and the Tow Foundation. G. M. was supported by Fondazione Malattie Miotoniche, Milan, Italy. Andrea Cortese would like to thank the Medical Research Council (MR/T001712/1), Cariplo Foundation, the Italian Ministry of Health (Ricerca Corrente 2018–2019), the Inherited Neuropathy Consortium and the Fondazione Regionale per la Ricerca Biomedica for the grant support.
Author information
Authors and Affiliations
Consortia
Contributions
UŠ and YA conducted experiments; U.Š., N.Š., A.-L.B. and M. Ro. analysed the data; U.Š. and E. Bur. designed the study; H. P. provided patient data collected by the NYGC ALS consortium; A.C., C.C., E. Bug., R.C., G.M., M.Ri., M.M. provided IBM patient samples; P.F., M.S. and E. Bur. supervised the study; U.Š. and E. Bur. wrote the manuscript with contributions of other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Gracjan Michlewski and Eve Rogers. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Šušnjar, U., Škrabar, N., Brown, AL. et al. Cell environment shapes TDP-43 function with implications in neuronal and muscle disease. Commun Biol 5, 314 (2022). https://doi.org/10.1038/s42003-022-03253-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-022-03253-8
This article is cited by
-
Inclusion body myositis, viral infections, and TDP-43: a narrative review
Clinical and Experimental Medicine (2024)
-
Modelling TDP-43 proteinopathy in Drosophila uncovers shared and neuron-specific targets across ALS and FTD relevant circuits
Acta Neuropathologica Communications (2023)
-
The era of cryptic exons: implications for ALS-FTD
Molecular Neurodegeneration (2023)
-
Cytotoxic immune cells do not affect TDP-43 and p62 sarcoplasmic aggregation but influence TDP-43 localisation
Scientific Reports (2023)
-
An impaired splicing program underlies differentiation defects in hSOD1G93A neural progenitor cells
Cellular and Molecular Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.