Abstract
Multi-resistant bacteria are a major threat in modern medicine. The gram-negative coccobacillus Acinetobacter baumannii currently leads the WHO list of pathogens in critical need for new therapeutic development. The maintenance of lipid asymmetry (MLA) protein complex is one of the core machineries that transport lipids from/to the outer membrane in gram-negative bacteria. It also contributes to broad-range antibiotic resistance in several pathogens, most prominently in A. baumannii. Nonetheless, the molecular details of its role in lipid transport has remained largely elusive. Here, we report the cryo-EM maps of the core MLA complex, MlaBDEF, from the pathogen A. baumannii, in the apo-, ATP- and ADP-bound states, revealing multiple lipid binding sites in the cytosolic and periplasmic side of the complex. Molecular dynamics simulations suggest their potential trajectory across the membrane. Collectively with the recently-reported structures of the E. coli orthologue, this data also allows us to propose a molecular mechanism of lipid transport by the MLA system.
Similar content being viewed by others
Introduction
Gram-negative bacteria are enveloped by two lipid bilayers, separated by the periplasmic space containing the peptidoglycan cell wall. This two-membrane system shields them effectively from a range of antibiotics like Penicillin, and also from chemicals like detergents or enzymes like lysozyme. Since many important bacterial pathogens like Pseudomonas aeruginosa, Campylobacter or Acinetobacter belong to this group of bacteria, they form an important target in modern medicinal research1. The two membranes in Gram-negative bacteria have distinct lipid compositions: the inner membrane consists of glycerophospholipids, with both leaflets having similar compositions, while the outer membrane is asymmetric, with an outer leaflet of lipopolysaccharides and an inner leaflet of glycerophospholipids2 (Fig. 1A). This lipid gradient, depicting the first and most important permeation barrier, is maintained by several machineries, including YebT, PqiB and the multicomponent maintenance of lipid asymmetry (MLA) system3,4, which consists of MlaA present in the outer membrane, the shuttle MlaC in the periplasmic space and the MlaBDEF ABC transporter system in the inner membrane (Fig. 1A). The structure of some of these components have previously been solved: the outer membrane protein MlaA, which was found to form a stable complex with outer membrane porins OmpF and OmpC5,6, and the periplasmic protein MlaC revealing a hydrophobic pocket for direct lipid transport through the periplasm3,7. Low-resolution cryo-EM maps of the MlaBDEF core complex, from Escherichia coli (MlaBDEFec)3 and Acinetobacter baumannii (MlaBDEFab)8 have also been reported, and revealed the overall architecture of the complex, but did not allow to elucidate the molecular details of lipid binding and transport. Opinions about the directionality of lipid transport by the MLA system have been highly controversial, with initial reports suggesting that it recycles lipids from the outer membrane to the inner membrane3,9,10, but recent results8,11,12 indicated that it might export glycerophospholipids to the outer membrane. Molecular dynamics simulations of the outer membrane MlaA, and periplasmic MlaC proteins have previously been reported in the literature6,7,11, but none of the MlaBDEF inner membrane complex.
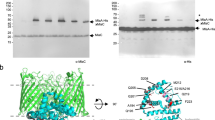
A Schematic representation of lipid transport by the MlaABCDEF system in Gram-negative bacteria (LPS = Lipopolysaccharide). B 3.9 Å Cryo-EM map of MlaBDEFab-AppNHp in β-DDM. C Cartoon representation of the MlaBDEFab atomic model. The complex has a global C2 symmetry, with six copies of MlaD that span the inner membrane from the periplasmic space to the cytosol, two copies of MlaE embedded in the membrane (yellow/gold), two copies of the ATPase MlaF in the cytosol (green), each bound to a copy of MlaB (blue/cyan). D Top view of the MlaBDEFab complex reveals C6 symmetry of MlaD. E C-terminal regions of MlaF bind the opposing MlaB subunit via a handshake mechanism.
In this study, we report the structure of the MlaBDEFab complex in detergent, in three nucleotide states, by single-particle cryo-EM. We also performed molecular dynamics simulations to gain insights into the dynamics of lipids within their observed binding sites. Collectively, this provides important insights into the mechanism of lipid transport by the MLA system, and about the characterization of membrane proteins in detergent.
Results
Structure of MlaBDEFab
We had previously reported the purification of MlaBDEFab in the presence of the detergent n-dodecyl β-D-maltoside (DDM), and its structure to ~ 8 Å, by single-particle cryo-EM, from data collected on a side-entry 200 kV microscope8. In order to improve the resolution of this structure, we collected a dataset of the same complex, in the presence of the non-hydrolizable ATP analogue App-NHp, using a state-of-the-art Titan Krios instrument. Using this better and larger dataset, we were able to refine the structure to ~3.9 Å resolution (Fig. 1B, Table 1, Supplementary Fig. 1 and Supplementary Data 1). This map allowed us to build a de novo atomic model using Rosetta13 (see ‘Methods’ for details).
As shown in Fig. 1B, C, the transmembrane multiprotein complex features a 6-fold symmetric assembly of MlaD, with the C-terminal helix forming a basket in the periplasmic space (Fig. 1D), and the N-terminal helix spanning the inner membrane (Fig. 1C). The N-terminal TM helices of MlaD are wrapped around the two MlaE molecules in the membrane, with three MlaD helices interacting asymmetrically with one MlaE, as reported previously8. Intriguingly, while MlaE was predicted to have 6 TM helices, we observe that TM1 does not traverse the membrane, but is monotropically embedded in the inner leaflet, a feature similar to the G5G8 human sterol exporter14, suggesting similar mechanisms between these complexes, and further supporting the MLA system as a lipid exporter. On the cytosolic side, MlaE is anchored into the ATPase MlaF via the coupling helix situated between TM3 and TM4 (Fig. 1), again similar to the G5G8 sterol exporter complex. MlaF is bound to MlaB away from the nucleotide binding site, similar to the recently reported E. coli MlaBF structure15, with the C-termini of MlaF binding the opposing MlaB subunit by a handshake mode (Fig. 1E). We note, however, that particle classification demonstrated that only ~50% of the particles included MlaB bound to both MlaF, leaving the other 50% bound to only one copy of MlaB (Supplementary Fig. 2). Dual binding of MlaB did not introduce major structural alterations at the detected resolution, and this observation may correspond to a regulatory role for MlaB or could be due to complex disassembly during sample preparation.
Lipids spontaneously bind into pockets of cytoplasmic MlaBDEFab
In the cryo-EM-derived map of MlaBDEFab, we observed well-defined density in the pocket formed between the MlaE TM1 and two MlaD helices, within the inner leaflet of the lipid bilayer (Fig. 2A, C), which could not be interpreted by protein atoms. We also observed that this pocket is coated by cationic residues, mainly Arg14, Arg47 and Arg234 of MlaE, forming a charged pocket that might attract a lipid head group (Fig. 2D). This observation prompted us to propose that this density corresponds to detergent molecules bound to the complex. As the sample was solubilized in DDM, this density is most likely occupied by the maltoside ring, with the flexible hydrophobic chains extending up into an apolar region of MlaE. In order to confirm if the observed density was consistent with lipid positioning, we performed two independent molecular dynamics simulations (Table 2) with unoccupied binding pockets as starting structures and observed rapid incorporation of bulk lipids during equilibration in both simulations (Fig. 2B, E, F). As an additional test, a further two simulations (Table 2) were performed by removing the lipids that had entered the binding sites, these were also run for 500 ns. Once again lipids rapidly moved to occupy the binding sites. Newly bound lipids remained stably bound thereafter during production runs (Fig. 2F) in all four simulations during 500 ns trajectories. The translational motion of these bound lipids was less than that of the ‘bulk’ lipids of the model inner membrane (Fig. 2F). In each of the four simulations, two lipids were observed entering the protein, one in each binding site. Interestingly we always observed one PE in one site, and PG in the other. Overall, each lipid within the binding site formed at least two hydrogen bonds with the protein over the course of the simulations. These hydrogen bonds were most often between the phosphate moiety of the lipid head group and arginine residues; R14, R108, R234, R365 and R491 (Supplementary Fig. 7). Other residues which participated in interactions with the lipids were W238 and W491, largely also through headgroup moieties. We propose here that lipids binding in these regions depicts a reasonable first step in lipid export.
A–C Transmembrane region of MlaBDEFab with one MlaE and three MlaD proteins shown. Red circles indicate density unoccupied by protein residues. Note that at this cut-off the detergent micelle is not visible, but the density attributed to the allocrit is clearly visible. D Electrostatic surface potential of modelled inner membrane MlaBDEFab (blue = +5 kT/e, white = 0 kT/e, red = −5 kT/e) shows a cationic binding site for anionic lipid head groups like DDM and an apolar pocket for the uncharged lipid tails. DDM was modelled inside the previously empty cryo-EM density (orange). E, F Molecular dynamics simulations show spontaneous, stable binding of lipids into the cytosolic binding pockets of MlaBDEF. Lipid-free MlaBDEFab was embedded into an inner membrane model system of 75% POPE, 20% POPG, 5% cardiolipin (headgroups as green spheres, see ‘Methods’ section for details). Panel (E) shows the trajectory of two POPE lipids during a 500 ns simulation, the colour scheme indicates the movement of the lipids as shown in the legend. In this simulation the lipids moved into this location spontaneously during the equilibration process, as shown in the close-up view in the inset in which the red coloured lipids indicate starting positions before the equilibration process. Panel (F) shows the motion of the lipids in the xy plane (inset) over 500 ns. The red points represent the centre of mass (in xy) of the lipids in the binding site, whereas the different shades of grey represent four ‘bulk’ membrane lipids from each simulation. Data from both independent simulations (r1 and r2) are shown at both temperatures. The data represents 100 frames taken at 5 ns intervals from each trajectory of 500 ns duration. The lipids are confined within an area of 10 × 10 Å for 500 ns, showing this location is favourable.
Lipids bind into the periplasmic basket and partially flip
The periplasmic region of MlaBDEFab consists mainly of hexameric MlaD forming a basket shape (Fig. 1). Similar to other MCE domain proteins16, MlaD consists of a central beta sheet motif with a central pore loop that is formed by hydrophobic Leu153/Leu154 in the centre of the C6 symmetric complex (Fig. 3A). We observed that in our map, unattributed density was present between the central pore loops, as well as in the central pore (Fig. 3A). Importantly, this density is not an artefact of the 6-fold symmetry, as it is also resolved when no symmetry was applied during the reconstruction (Supplementary Fig. 8). The presence of lipid molecules in very similar positions were previously reported in the MCE protein YebT17, which prompted us to postulate that this unattributed density on the MlaD periplasmic region also corresponds to detergent molecules. The basket region has previously been proposed to form the binding site for the periplasmic carrier protein MlaC5, suggesting that lipids are extracted from this position upon MlaC binding, consistent with the interpretation that these regions of the map correspond to lipid molecules.
A Close-up of the periplasmic MlaD basket that houses six peripheral and one central detergent molecule. B–D Spontaneous flipping of the centrally bound lipid within the basket region of MlaBDEFab during 250 ns molecular dynamics simulations. Panel (B) shows the centre of mass motion of the seven lipids in the xy plane. They are confined to an area of ~5 × 5 Å, indicating this is a high lipid affinity region. Panel (C) shows the centre of mass movement of the seven lipids in the z dimension as a function of time. The lipid shown in panel (A) corresponds to the black curve showing a clear movement towards the cytoplasmic end. Three other lipids (blue) move into this channel but to a lesser extent than the aforementioned lipid, whereas three others (orange) remain close to their starting positions. Simulations from a model at the reported resolution cannot clarify the directionality of the movement, but rather that these regions are conduits for lipids. Panel (D) shows a cutaway view of the protein with a POPE lipid at two time points during the simulation, time = 0 ns and 150 ns. The simulation was initiated with seven POPE lipids placed at the periplasmic end of the protein corresponding to the density for detergents in the cryo-EM data (Supplementary Table 2). The lipid which is displaced more towards the cytoplasmic end is from the frame at t = 150 ns. The central hydrophobic ‘channel’ of the protein is a clear conduit for lipids given the spontaneous movement of POPE into this region in just 150 ns.
Because of this likely important role of lipid dynamics for this region, we next performed 3D variability analysis18, to identify molecular motions in the MlaD basket. As shown in Supplementary Movies 1 and 2, this analysis revealed that the 6x MlaD basket can be translated and rotated against the MlaBEF transmembrane part, which may play a role in the lipid transport mechanism. Incidentally, this dynamic property likely also limits the achievable resolution for this region of the map.
To further investigate the dynamic properties of the lipids present in the MlaD basket, we set up two systems, one in which we replaced the DDM molecules with 7x PE lipids and one in which we replaced the DDM molecules with 4x PE and 3x PG lipids, and subjected both systems to molecular dynamics simulations (Table 2). PE and PG were chosen here, given they have both been shown to bind to MlaBDEF (E. coli) by mass spectrometry19 and thin-layer chromatography10. As shown in Fig. 3, we observed that the peripheral lipids are quite stable within the basket during the course of the simulation. In contrast, the central lipid undergoes a motion within 150 ns of the trajectory. Remarkably, during this motion, we observed lipid flipping (Fig. 3D), with the head group which was modelled away from the central pore operating at an almost 180°, with the polar group buried within the MlaE transporter. This observation likely indicates that we had initially built the lipid in the wrong orientation, and highlights the importance of properly modelling lipid molecules, especially at the intermediate resolution such as that of our MlaBDEFab map. In addition, this demonstrates the presence of a large hydrophobic pocket at the MlaD–MlaE interface, which could correspond to the channel for lipid transport.
Nucleotide binding occurs at the interface of MlaE, MlaF and MlaD
As indicated above, our structure of MlaBDEFab was determined in the presence of the non-hydrolizable ATP analogue AppNHp. Accordingly, we observed clear density of the nucleotide and magnesium, within the MlaF binding pocket (Fig. 4A). The nucleotide links the ATPase subunits with the lipid transport domains at the interface of MlaF, MlaE and the N-terminus of MlaD. The resolution was sufficient to map the phosphate binding regions like Walker-A motif around Lys55 of MlaF, as well as binding of the γ-phosphate by Ser51 and His211 (H-loop). The Mg2+ ion is coordinated by the Walker-B motif around Asp177 (Fig. 4A) and adenosine ring binding is achieved through Arg26 in the A-loop. As revealed in the previously reported low-resolution map8, the position of MlaF suggest that the complex is in the substrate-bound conformation.
A AppNHp is bound at the interface of MlaE (orange), MlaD (red) and MlaF (green). B maps of MlaBDEFab bound to AppNHp (grey), its apo state (yellow) and bound to ADP (cyan). C The overlay of the maps unambiguously confirms the presence of nucleotide in the AppNHp and ADP maps, but not in the apo map. Nonetheless, no overall structural changes are observed.
In order to identify structural changes in the complex associated with ATP binding and hydrolysis, we next determined the structure of MlaBDEFab without nucleotide (Supplementary Fig. 3), and with ADP (Supplementary Fig. 4). As shown in Fig. 4A–C, we obtained both structures, at ~4.2 and ~ 4.4 Å, respectively (Table 1 and Supplementary Data 2 and 3). We note that in spite of the nominal global resolution, the map for the complex bound to ADP possesses features largely similar to that of the complex bound to AppNHp. In contrast, the map of the apo complex is less well-resolved, in particular with most TM helices being mostly featureless. This suggests that nucleotide binding stabilizes the overall architecture of the MlaBDEFab complex.
Nonetheless, we note that the overlay of the MlaBDEFab maps in the apo, AppNHp-bound and ADP-bound states shows no major structural changes in any of the complex components, other than the nucleotide binding site (Fig. 4D). This indicates that in spite of the nucleotides being bound to the ATPase domain, the conditions used here are not sufficient to trigger the activation of the channel opening.
Discussion
While this manuscript was under review, three independent groups released the structure of the E. coli MlaBDEF complex (MlaBDEFec), in a range of nucleotide states and with various solubilization approaches19,20,21.
Comparison of the MlaBDEFec structure (PDB-ID 7CH0, 7CGN), determined in lipid nanodiscs, and the MlaBDEFab structure (this study), determined in DDM, reveals an ~10 Ang movement of the MlaD TM helices, accompanied by an ~30° tilt of the MlaE N-terminal helix (Fig. 5A). This movement narrows the lipid binding pocket (Fig. 5). A structural flexibility like this would allow adjustment of MlaBDEF for various lipids. However, as lateral pressure in the membrane plane of a detergent micelle is not comparable to a lipid nanodisc, differences in sample preparation can also cause movement of this hinge. Furthermore, although the MLA system is conserved in Gram-negative bacteria, the sequence identity between the MlaBDEFab and MlaBDEFec proteins is around 30–40% identical (depending on the protein, Supplementary Fig. 9), and it is therefore possible that the differences observed between the two structures correspond to variability between bacterial species.
The cryo-EM map of MlaBDEFec in lipid nanodiscs19 and of MLABDEDab in a DDM micelle (this study) show large differences in the lower lipid binding sites. A Both the MlaD N-terminal helix and the MlaE N-terminal helix form a much smaller pocket (red circle) in detergent environment (yellow, this study) compared to lipid environment (grey, EMD-30355). Two states were observed in the MlaBDEFec study; tall (B) and close (C). Comparison between MlaBDEFec and MlaBDEFab shows that the pocket is smaller in both conformations, whereas the nucleotide position is identical in all three structures.
We note that, in one of the aforementioned studies, two ATP-bound conformational states were observed for MlaBDEFec: a more open conformation (tall state) and a tightly bound conformation (close state), with a shifted nucleotide position. The nucleotide position in MlaBDEFab in detergent resembles the tall state with an open conformation of MlaF (Fig. 5B, C). Our variant analysis did not reveal these two conformations in MlaBDEFab, which might therefore be due to the different orthologues, or the different method for solubilization.
We also emphasize that the detergent binding pockets observed in our MlaBDEFab structures are also confirmed in the MlaBDEFec structures, notably the lipids found between the MlaD TM and the N-terminal helix of MlaE19, and the detergent present in the central cavity19,20,21. For the latter, different exact detergent positions are observed depending on the state and study, but they are largely consistent with the lipid positions we have obtained in the MD simulations, with the charged group buried at the MlaE–MlaD interface. However, our map is the only one with clear detergent density at the interface between MlaD molecules in the periplasmic site. As mentioned above, this could correspond to species specificity, or could be an artefact of the use of detergent for structure determination.
In the light of the multiple structures now available, the structures of MlaBDEFab reported here support a mechanism for lipid export as summarized in Fig. 6. We observe lipids binding to the inner lipid binding pocket; this region is likely flexible and can possibly adapt to different lipids. MlaE is in very close proximity to ATP in the closed conformation, possibly describing an additional regulatory mechanism in A. baumannii. Structures of MlaBDEFec also showed a large-scale structural alteration upon tight ATP binding19 that might be restrained due to the detergent environment in our experiments. We could furthermore observe a central lipid binding site in the periplasmic basket region of MlaD that partially flips into the central channel of MlaE during the course of our MD simulation. Nonetheless, neither our structures of MlaBDEFab nor the aforementioned MlaBDEFec could resolve lipid transition between the cytoplasmic lipid binding pocket and the central channel. Similarly, lipid transport from MlaBDEF to MlaC remains elusive. These open questions are also difficult to address with molecular simulations, and it is useful here to reflect on the wider utilities and limitations of MD for studying a system of the size and complexity of the MlaBDEF complex. The utilities of standard, equilibrium MD are clear; the movement of lipids in and around the protein and protein conformational dynamics (within the timescale limitations of MD) can be probed, as we have done here. Any preference for one lipid type over another for being transported by the Mla system is more difficult to determine using standard MD due to limited sampling. Here, free-energy methods are more useful such as those demonstrated by Corey et al. with coarse-grained (CG) force fields22. Coarse-grained models also provide a more efficient route to determine more long-range effects of the protein on the lipid environment such as membrane thinning/widening or lipid sorting. These aspects are not the focus of the current study, but will be probed in future work.
A Structural model, and (B) observed degrees of freedom in this study. Lipids can move freely into the cytosolic paired binding pockets (‘entry/exit’) that are very close to the ATP binding sites that directly bind MlaD (‘lock’). Lipids can move and even flip inside the basket region (‘transfer’). The elucidated structures could not show how transport from the entry sites to the basket region is performed and how lipid exchange to/from MlaC is achieved.
It should be emphasized that the structural studies reported here favour an anteretrograde direction for lipid transport by structure similarity, as supported by our previous biochemical data in A. baumannii21 as well as other studies on E. coli12,19,21. Nonetheless, other assays point towards a retrograde directionality20,23, and the current structures do not conclusively preclude this. Further structural analyses, in particular in complex with the periplasmic carrier MlaC, will be required to resolve this controversy definitively.
Methods
MlaBDEFab protein expression and purification
The expression and purification of MlaBDEFab has been described previously8. Briefly, the plasmid encoding the full operon under the control of a T7 promoter was transformed into E. coli BL21 DE3 cells and grown at 37 °C until the cell density reached OD (600 nm) = 1.0. The temperature was reduced to 20 °C before induction with 1 mM isopropyl β-D-thiogalactoside (IPTG) and incubation overnight. Cells were harvested using centrifugation at 5000g and resuspended in ice-cold buffer A (20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5% (v/v) glycerol) before disruption in an ultrasonicator on ice (6 cycles; 60 s run, 30 s cool down). Cell debris were pelleted at 17,000g for 10 min, and the membrane fraction was separated by centrifugation at 100,000g for 1 h. This pellet was resuspended by gentle stirring in Buffer A supplemented with 1% (w/v) dodecyl-β-d-maltopyranoside (DDM) at 7 °C for 1 h. After another centrifugation step at 100,000g for 30 min the supernatant was applied to a 5 ml Ni-NTA superflow column (GE Healthsciences) equilibrated with buffer A supplemented with 20 mM imidazole and 0.025% (w/v) DDM. The column was washed with buffer A supplemented with 20 mM imidazole and 0.025% (w/v) DDM before elution with buffer A supplemented with 300 mM imidazole and 0.025% (w/v) DDM was performed. The elute was concentrated to 5 ml and ran through a 16-600 HiLoad Superdex 200 pg gel filtration column (GE Healthcare), preequilibrated with 20 mM Hepes (pH 7.0), 150 mM NaCl, 0.025% (w/v) DDM. Peak fractions were collected, purest fractions were selected using SDS-PAGE and concentrated to 5 mg/ml.
Cryo-EM sample preparation, data acquisition and image processing
In all, 4 µl freshly purified 5 mg/ml MlaBDEFab in 20 mM Hepes (pH 7.0), 150 mM NaCl, 0.025% (w/v) DDM was applied on freshly glow-discharged 300 mesh Quantifoil R2/2 grids, blotted for 3.5 s in a Leica EM-GP plunge freezer at 80% humidity and 4 °C, before getting plunged into liquid ethane at −170 °C. For investigation of the AppNHp and ADP states, the protein was mixed 1:1 with 20 mM of the corresponding nucleotide in 50 mM Hepes (pH 8.0), 150 mM NaCl, 0.025% (w/v) DDM buffer and incubated for 60 min on ice prior to grid preparation.
Micrographs of MlaBDEFab-AppNHp were recorded on a 300 kV Titan Krios microscope with a Gatan K2 Summit detector in counting mode. In all, 2557 movies were recorded with a pixel size of 1.07 Ang in 47 frames with 1 e−/Ang2/frame. In all, 1901 movies of MlaBDEFab-ADP were recorded on the same instrument with a total dose of 40 e−/Ang2. The MlaBDEFab apo dataset was recorded on a 300 kV Titan Krios equipped with a Gatan K3 bioquantum detector. In all, 4737 micrographs with a pixel size of 0.41 Ang were recorded with a total dose of 40 e−/Ang2 in 50 frames. Data processing was performed in CryoSPARC v2.14.2 (see Supplementary Figs. 1 and 3 for details).
Model building
The atomic model of MlaBDEFab was manually built into the high-resolution regions of the MlaBDEFab-AppNHp map using Coot. Computational intervention was required to build residues 4-38 and 94-134 of MlaD, as well as refine the AppNHp binding site of MlaF. RosettaES13 was used to model residues 4-38 for each of the 6 MlaD subunits using the manually built model as the starting point. Residues 94-134 were first modelled with Rosetta ab initio24, which yielded a tightly converged ensemble with a 2-helix topology. The top scoring model from the ab initio predictions was unambiguously docked into the corresponding density for each of the MlaD subunits using UCSF Chimera25. Loops were completed and the entire MlaD basket refined using RosettaCM26 with the context of the cryo-EM density. To refine the AppNHp binding site of MlaF, we first used RosettaCM to hybridize the manually built model with the homologous structures of (3fvq, chain A), and (4ki0, chain B) in the cryo-EM density. Then using homology models and the unexplained density as a guide, a modified version of the Rosetta protocol GALigandDock (unpublished) that uses the cryo-EM data to drive sampling was used to dock the AppNHp molecule. As input to the protocol, a mol2 file of AppNHp was modified using Open Babel (v. 2.4.1)27 to add hydrogens, charges were assigned with the AM1-BCC charge method in antechamber28 and a params file was generated with the script main/source/scripts/python/public/generic_potential/mol2genparams.py, which is distributed with Rosetta. Finally, the entire complex was refined using RosettaCM, and magnesium atoms were added by incorporating distance and angle constraints between the Mg atoms and the AppNHp oxygens on the β and γ phosphates during the final minimization. ISOLDE in ChimeraX was used to manually correct for modelling errors. Phenix.real.space.refine was used for refinement and validation. As shown in Supplementary Figs. 5 and 6, the resulting model has an excellent fit to the map density, with clear fit of the side-chains in particular for the TM helices of MlaE and MlaD, which were critical for determining their registry. We note nonetheless that the TM for one of the MlaD molecules is mostly featureless (orange in Supplementary Fig. 6), in which case we relied on the other two copies to position the helix in the density.
Molecular dynamics system preparation
The MlaBDEFab structure was completed by adding the missing residues using Modeller 9.23 (http://salilab.org/modeller/)29,30. The completed protein structure was embedded in an inner E. coli inner membrane with dimensions of ~21 × 21 × 18 nm using the CHARMM GUI web server31,32,33,34,35,36,37. We used a simplified version of the A. baumannii inner membrane composition38: 75% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 20% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) and 5% 1′,3′-bis[1,2-dioleoyl-sn-glycerol-3-phospho-]-sn-glycerol, cardiolipin (CDL). To relax any steric conflicts within the system generated during set up, energy minimization of 5000 steps was performed on the starting conformation using the steepest descent method. An equilibration procedure followed in which the protein was subjected to position restraints with different force constants. The full equilibration protocol is shown in Supplementary Table 1. The Mla_323 system was set up by removal of lipids that had entered the proposed binding sites during equilibration. An additional set of simulations, Mla_7PE and Mla_4PE3PG were set up in which 7 lipid molecules (PE or PE + PG, details are in Table 1) were positioned such that the lipid head group was overlaid on the maltose moiety of the DDM molecules and the lipid tails were aligned along the DDM hydrocarbon chain using visual molecular dynamics (VMD)39.
Two independent simulations of each system were performed (denoted r1 and r2 in the main text). The simulation cell had dimensions of 210 × 210 × 180 Å, thus the membrane patch had dimensions of 210 × 210 Å.
Equilibrium molecular dynamics simulations
All simulations were carried out with GROMACS 2019.640 version (www.gromacs.org) and CHARMM36 forcefield41. For the nonbonded interactions and the short-range electrostatics cut-offs of 1.2 nm was applied to the system with the potential shift Verlet cut-off scheme, whereas the long-range electrostatic interactions were treated using the particle mesh Ewald (PME) method42. All atoms were constrained using the LINCS algorithm43 to allow a time step of 2 fs. The desired temperature of either 310 or 323 K was controlled with Nose-Hoover thermostat44,45 (1.0 ps coupling constant). The pressure was maintained at 1 bar using the Parrinello-Rahman46 semi-isotropic barostat with a coupling constant of 1.0 ps. Equilibrium MD systems were performed with two repeats per system, where each repeat was run with different initial velocity seeds. The summary of all production runs is shown in Supplementary Table 2. The results were analysed using GROMACS tools. Molecular graphics were generated using VMD.
Note added in proof
Since this paper was submitted to Communications Biology, an independent study reported the structure of the MlaBDEFab complex, in nanodisc, in the absence of nucleotide47. This paper supports the conclusions of this study.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Source data underlying figures are presented in Supplementary Data 1–3. Raw images of the MlaBDEFab-AppNHp dataset are publicly available under EMPIAR-10425. Sharpened maps in C2, C1 and C6 symmetry, masks and half-maps of MlaBDEFab-AppNHp, ADP and apo are publicly available under EMD-11082, EMD-11083 and EMD-11084, respectively. The MlaBDEFab-AppNHp atomic model is publicly available under PDB-6Z5U. All other data are available from the corresponding author on reasonable request.
References
Bernier, S.P., Son, S. & Surette, M.G. The Mla pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components.J. Bacteriol. 200, e00156-18 (2018).
Zhang, Y.-M. & Rock, C. O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6, 222–233 (2008).
Ekiert, D. C. et al. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169, 273–285.e17 (2017).
Isom, G. L. et al. LetB structure reveals a tunnel for lipid transport across the bacterial envelope. Cell 181, 653–664.e19 (2020).
Abellón-Ruiz, J. et al. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat. Microbiol. 2, 1616–1623 (2017).
Yeow, J. et al. The architecture of the OmpC–MlaA complex sheds light on the maintenance of outer membrane lipid asymmetry in Escherichia coli. J. Biol. Chem. 293, 11325–11340 (2018).
Huang, Y.-M. M. et al. Molecular dynamic study of MlaC protein in Gram-negative bacteria: conformational flexibility, solvent effect and protein-phospholipid binding. Protein Sci. 25, 1430–1437 (2016).
Kamischke, C. et al. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. eLife 8, e40171 (2019).
Malinverni, J. C. & Silhavy, T. J. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl Acad. Sci. USA 106, 8009–8014 (2009).
Thong, S. et al. Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. eLife 5, e19042 (2016).
Hughes, G. W. et al. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat. Microbiol. 4, 1692–1705 (2019).
Hughes, G. W., et al. MlaFEDB displays flippase activity to promote phospholipid transport towards the outer membrane of Gram-negative bacteria. Preprint at https://www.biorxiv.org/content/10.1101/2020.06.06.138008v1.full (2020).
Frenz, B., Walls, A. C., Egelman, E. H., Veesler, D. & DiMaio, F. RosettaES: a sampling strategy enabling automated interpretation of difficult cryo-EM maps. Nat. Methods 14, 797–800 (2017).
Lee, J.-Y. et al. Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature 533, 561–564 (2016).
Kolich, L.R. et al. Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation.eLife 9, e60030 (2020).
Isom, G. L. et al. MCE domain proteins: conserved inner membrane lipid-binding proteins required for outer membrane homeostasis. Sci. Rep. 7, 1–12 (2017).
Liu, C., Ma, J., Wang, J., Wang, H. & Zhang, L. Cryo-EM structure of a bacterial lipid transporter YebT. J. Mol. Biol. 432, 1008–1019 (2020).
Punjani, A. & Fleet, D. J. 3D variability analysis: directly resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM images. J. Struct. Biol. 213, 107702 https://www.sciencedirect.com/science/article/pii/S1047847721000071 (2021).
Chi, X. et al. Structural mechanism of phospholipids translocation by MlaFEDB complex.Cell Res. 30, 1127–1135 (2020).
Tang, X. et al. Structural insight into outer membrane asymmetry maintenance of Gram-negative bacteria by the phospholipid transporter MlaFEDB.Nat. Struct. Mol. Biol. 28, 81–91 (2021).
Coudray, N. et al. Structure of bacterial phospholipid transporter MlaFEDB with substrate bound.eLife 9, e62518 (2020).
Corey, R. A., Vickery, O. N., Sansom, M. S. P. & Stansfeld, P. J. Insights into membrane protein–lipid interactions from free energy calculations. J. Chem. Theory Comput. 15, 5727–5736 (2019).
Powers, M. J., Simpson, B. W. & Trent, M. S. The Mla pathway in Acinetobacter baumannii has no demonstrable role in anterograde lipid transport. eLife 9, e56571 (2020).
Simons, K. T., Bonneau, R., Ruczinski, I. & Baker, D. Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins 37, 171–176 (1999).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Song, Y. et al. High resolution comparative modeling with RosettaCM.Structure 21, 1735–1742 (2013).
O’Boyle, N. M. et al. Open Babel: an open chemical toolbox. J. Cheminform. 3, 33 (2011).
Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25, 247–260 (2006).
Šali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Sánchez, R. and Šali, A. Comparative protein structure modeling: introduction and practical examples with modeller. In Protein Structure Prediction (ed. Webster, D. M.) 97–129 (Humana Press, 2000).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Brooks, B. R. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009).
Lee, J. et al. CHARMM-GUI input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Wu, E. L. et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014).
Jo, S., Lim, J. B., Klauda, J. B. & Im, W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 (2009).
Jo, S., Kim, T. & Im, W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE 2, e880 (2007).
Lee, J. et al. CHARMM-GUI membrane builder for complex biological membrane simulations with glycolipids and lipoglycans. J. Chem. Theory Comput. 15, 775–786 (2019).
Lopalco, P., Stahl, J., Annese, C., Averhoff, B. & Corcelli, A. Identification of unique cardiolipin and monolysocardiolipin species in Acinetobacter baumannii. Sci. Rep. 7, 2972 (2017).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–28 (1996). 33–8.
Van Der Spoel, D. et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005).
Huang, J. & MacKerell, A. D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Hoover, W.G. Canonical dynamics: equilibrium phase-space distributions.Phys. Rev. A 31, 1695–1697 (1985).
Nosé, S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 52, 255–268 (1984).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Zhang, Y., Fan, Q., Chi, X., Zhou, Q. & Li, Y. Cryo-EM structures of Acinetobacter baumannii glycerophospholipid transporter.Cell Discov. 6, 86 (2020).
Acknowledgements
We are thankful for financial support through BBSRC project BB/R019061/1. We acknowledge Diamond Light Source for access and support of the cryo-EM facilities at the UK’s national Electron Bio-imaging Centre (eBIC) (under proposal EM-19832). We thank Emma Hasketh and Rebecca Thompson from the Astbury Centre for Structural Molecular Biology Leeds for support during measurements. We acknowledge support from LonCEM, King’s College London during measurements. The University of Sheffield FoS cryo-EM facility was used for grid preparation and optimization. We are grateful to Justin Kollman for help with the initial stages of this project. We acknowledge the use of the ARCHER supercomputer via HECBioSim, funded through EPSRC project EP/R029407/1, and the use of the IRIDIS High Performance Computing Facility at the University of Southampton.
Author information
Authors and Affiliations
Contributions
D.M. purified the MlaBDEF complex, performed the cryo-EM experiments, and processed the data, with advice from J.R.C.B.; S.B.T. assisted with cryo-EM grid screening. D.M., D.P.F. and A.M. built the atomic model, with support from J.R.C.B. and F.M.; K.S. and S.K. performed the MD simulation experiments. J.F. made the expression construct and developed the MlaBDEFab purification protocol, and helped with purification, with support from S.I.M. D.M. and J.R.C.B. wrote the manuscript, with comments from all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: George Inglis. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mann, D., Fan, J., Somboon, K. et al. Structure and lipid dynamics in the maintenance of lipid asymmetry inner membrane complex of A. baumannii. Commun Biol 4, 817 (2021). https://doi.org/10.1038/s42003-021-02318-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-021-02318-4
This article is cited by
-
An octameric PqiC toroid stabilises the outer-membrane interaction of the PqiABC transport system
EMBO Reports (2024)
-
Molecular mechanism of phospholipid transport at the bacterial outer membrane interface
Nature Communications (2023)
-
The Mla system of diderm Firmicute Veillonella parvula reveals an ancestral transenvelope bridge for phospholipid trafficking
Nature Communications (2023)
-
Comparative insight into the effects of different carbon source supplement on antibiotic resistance genes during whole-run and short-cut nitrification-denitrification processes
Environmental Science and Pollution Research (2023)
-
Polymyxin B1 within the E. coli cell envelope: insights from molecular dynamics simulations
Biophysical Reviews (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.