Abstract
Pathogen-induced cancers account for 15% of human tumors and are a growing concern for endangered wildlife. Fibropapillomatosis is an expanding virally and environmentally co-induced sea turtle tumor epizootic. Chelonid herpesvirus 5 (ChHV5) is implicated as a causative virus, but its transmission method and specific role in oncogenesis and progression is unclear. We applied environmental (e)DNA-based viral monitoring to assess viral shedding as a direct means of transmission, and the relationship between tumor burden, surgical resection and ChHV5 shedding. To elucidate the abundance and transcriptional status of ChHV5 across early, established, regrowth and internal tumors we conducted genomics and transcriptomics. We determined that ChHV5 is shed into the water column, representing a likely transmission route, and revealed novel temporal shedding dynamics and tumor burden correlations. ChHV5 was more abundant in the water column than in marine leeches. We also revealed that ChHV5 is latent in fibropapillomatosis, including early stage, regrowth and internal tumors; higher viral transcription is not indicative of poor patient outcome, and high ChHV5 loads predominantly arise from latent virus. These results expand our knowledge of the cellular and shedding dynamics of ChHV5 and can provide insights into temporal transmission dynamics and viral oncogenesis not readily investigable in tumors of terrestrial species.
Similar content being viewed by others
Introduction
Sea turtle fibropapillomatosis (FP) is an epizootic (animal epidemic) tumor disease, affecting endangered sea turtles worldwide1,2,3,4,5. The disease is characterized by the formation of cutaneous and internal fibro-epithelial tumors, which can lead to debilitation and death. Fibropapillomatosis of sea turtles continues to spread geographically, and is now present in every major ocean basin in which green sea turtles (Chelonia mydas) are endemic (www.cabi.org/isc/datasheet/82638)3,6,7,8,9,10,11,12,13,14,15,16. Rates of FP are also increasing in many long-term affected locations, with incidence of the disease being the highest in near-shore habitats3,9,17,18,19,20,21. The FP-afflicted turtles studied here were found stranded in northeastern Florida. Fibropapillomatosis only began affecting C. mydas in northern Florida in the last decade, despite being present in areas of southern Florida since at least the early 1900s5,12,19,22,23,24. Concurrent with geographic expansion of the disease, incidence of FP in stranded C. mydas across the state of Florida has risen from 13.3% in 2005 to 42% in 20165,9,17,19.
Of all sea turtle species and life-stages, juvenile green sea turtles are most severely afflicted by FP. Long-lived reptiles have normally robust anti-cancer defenses, and with the exception of FP, reports of neoplasia in sea turtles are rare25,26,27,28,29. However, nearshore environmental exposures likely impair tumor suppressor mechanisms in sea turtles, thereby enabling virally-induced tumorigenesis3,30. While some environmental co-factors, such as eutrophication have been studied, their role in the pathogenesis of FP remains experimentally unconfirmed. It is possible that environmental exposures induce immunosuppression in sea turtles, thereby enabling viral loads to increase to the point of crossing an oncogenic threshold, similar to a number of human virally-induced cancers3,5,31,32,33. However, the pathogenesis of FP remains elusive. A chelonian-specific alphaherpesvirus (chelonid herpesvirus 5, ChHV5) has been implicated as a cause. However, Koch’s postulates to confirm its causative role have yet to be fulfilled, because ChHV5 is extremely difficult to isolate and propagate in the laboratory3,5,34,35,36,37. Similarly, despite advances in FP tumor research1,3,5,30,31,38,39,40,41,42,43,44,45,46, many open questions remain regarding the role of ChHV5 in driving FP tumorigenesis, including whether it is a cause of the disease or an opportunistic pathogen, exploiting immunocompromised tumor-afflicted turtles3,5,31. Interestingly, levels of ChHV5 (ChHV5 gB and UL30 gene DNA detected by qPCR, and ChHV5 glycoprotein H peptides detected by ELISA) in clinically healthy turtles are closer to that of FP tumors, than in non-tumored tissue of FP-afflicted turtles34,47. It is also unclear whether ChHV5 is lytic or latent in FP tumors and the occurrence of viral shedding and transmission is not well understood3,5,48,49,50,51.
In many locations, such as Florida, live sea turtles with FP that are found stranded are admitted to rehabilitation facilities for treatment, which often includes tumor resection surgery. Sea turtles under treatment for FP provide a valuable opportunity to study this disease in a manner that is infeasible or impractical in free-ranging animals. The combination of accessible rehabilitating patients and modern transcriptomics and genomics of different tumor types can address the open questions relating to ChHV5’s oncogenic role, while novel environmental DNA (eDNA) approaches can help resolve the dynamics of viral shedding and transmission.
Environmental DNA is a non-invasive forensics approach to the extraction and identification of organismal DNA fragments (genetic material) released into the environment, and this rapidly advancing approach is capable of improving endangered species detection and early pathogen detection52,53,54,55,56,57,58,59,60,61. Environmental samples can be analyzed for micro- and macro-organisms by several eDNA methods including metabarcoding and species-specific quantitative PCR (qPCR)55,57,62. The development of a rapid and high-throughput sampling scheme to detect virus shedding into the marine environment would benefit pathogen surveillance efforts, and consequently the performance of wildlife health status monitoring could be improved48,63. Here we applied qPCR and shotgun sequencing (“unbiased”, non-barcoded) eDNA approaches to temporally quantify ChHV5 viral shedding from rehabilitating patients. Such novel approaches are a particularly beneficial feature of aquatic models of virally-induced tumors, and will enable greatly improved understanding of the dynamic relationship between viral load and viral shedding, which is not readily measurable in terrestrial species.
We demonstrated previously that ChHV5 was transcriptionally latent in a small cohort of seven established external FP tumors5. We postulated that ChHV5 might be latent in established tumors, but more active during crucial early stage tumor initiation events, akin to the ‘hit and run’ hypothesis of viral oncogenesis5. To investigate this hypothesis, we employed deep sequencing-based transcriptomics and whole-genome sequencing (WGS) to determine the viral load and transcriptional status of ChHV5 across a variety of FP tumor stages and presentations: new external, established external, post-surgical regrowth external, internal lung, internal urinary bladder, and internal kidney tumors. Analysis of the host aspects of the current study’s transcriptomic and genomic data are explored in a companion paper38. Here we investigate viral dynamics during FP tumor growth, post-surgical recurrence, and correlations between ChHV5 viral load and ChHV5 gene expression with patient rehabilitation outcome to identify characteristics relevant to disease severity and fate of turtles afflicted with FP. Furthermore, we investigated potential routes of transmission, primarily horizontal transmission by direct shedding of ChHV5 into the water column, but also confirming the potential role of intermediary vectors and raising the possibility of vertical transmission. Fibropapillomatosis genomics29,31 and eDNA-based pathogen monitoring can reveal the precise mechanisms through which the virus is transmitted and the role of ChHV5 in host cell transformation and tumor progression. Such research will provide insights into this wildlife epizootic and reveal how ChHV5 can rapidly induce novel cancer incidence on an epidemic scale. Such information is vital to enable improved management, treatment, and mitigation strategies to be developed to combat this sea turtle conservation-relevant disease epizootic.
Results
High prevalence of ChHV5 in marine leeches feeding on FP-afflicted turtles
A number of potential routes for ChHV5 transmission between sea turtles have been postulated, including via vectors such as Ozobranchus leeches51. These marine leeches are commonly found on FP-afflicted turtles, often at high density within the crevices of external FP tumors (Fig. 1a, b). In line with previous studies51, we confirmed that ChHV5 could specifically be detected from DNA extracted from leeches that had fed on FP-afflicted turtles (Fig. 1c–e). All leech pooled samples removed from FP tumors tested positive for ChHV5 DNA (Fig. 1c, d), while half of the leeches removed from non-tumor locations of FP-afflicted animals were ChHV5 positive (Fig. 1c). Of 30 leeches from FP tumors assessed individually (one DNA extraction per whole leech), 90% were positive for ChHV5 (Fig. 1e). The 3 leeches that tested negative for ChHV5 (Fig. 1e) had smaller (barely visible) blood pellets than the other 27 leeches. Leeches removed from FP-free animals did not test positive for ChHV5 (Fig. 1c).
a Green sea turtle inguinal external FP tumor, infested with leeches. Upon patient intake FP-afflicted tumors frequently harbor marine leeches, as was the case for this patient 02-2021-Cm “Broccoli”. Leeches from “Broccoli’s” tumors were used for the ChHV5 analysis in (e). Leeches are commonly found within the crevasses of external FP tumors (right image). b Detailed view of a marine leech removed from the surface of a fibropapillomatosis tumor, with gills and with dark red blood pellet (after feeding on a C. mydas turtle) visible. c Detection and quantification of ChHV5 UL30 gene DNA by qPCR, using leeches as proxy eDNA samples (whole leech lysis and DNA extraction). Error bars denote the standard deviation of three technical replicates. Amplification ratio for leeches from the FP-free loggerhead turtle (09-2015-Cc) was 0, and the amplification ratio for both the FP-tumor leech and FP-tumor tissue samples (green turtle, 07-2015-Cm) was 1.0. d Quantification of ChHV5 UL30 gene DNA by qPCR, from leeches removed from FP-afflicted green turtles from either FP tumors or non-tumor locations. Individual turtle denoted by rd—36-2020-Cm “Richard Dawkins”, bh—52-2020-Cm “Bruno Hofer”, or rg—78-2020-Cm “Ruth Gates”. Approximately ten leeches were pooled for each of the 12 DNA extraction samples. Error bars denote the standard deviation of six technical replicates. e Quantification of ChHV5 UL30 gene DNA by qPCR, from leeches (individual leech DNA extractions) removed from FP tumors of green sea turtle patient 02-2021-Cm “Broccoli” (a). Amplification ratios for leech samples are provided in Supplementary Table 1. Error bars denote the standard deviation of six technical replicates.
Detection of ChHV5 shedding into the water column by environmental DNA (eDNA) approaches reveals novel shedding dynamics and tumor burden correlations
Another potential mode of transmission is direct ChHV5 shedding into the environment. Direct viral shedding as a route of transmission is thus far only supported by indirect evidence, i.e., cloacal swabs, urine, ocular, oral, and nasal secretions49,50,64, viral inclusion bodies near the surface of external tumors48 and the elevated levels of ChHV5 detected in bladder tissue (see below). Direct detection of ChHV5 in the water column is lacking. Therefore, we employed environmental DNA (eDNA)-based approaches coupled with qPCR and next-generation sequencing to detect ChHV5 in patient tank water. Such novel approaches can aid in answering previously intractable questions about direct ChHV5 transmission, such as presence, abundance, and persistence in the marine environment. First, we determined that the presence of ChHV5 could be readily detected in eDNA extracted from tank sea water, using either an established37 UL30 qPCR assay (Fig. 2a) or next-generation sequencing (see WGS section below). ChHV5 was detectible not only from patient tank water, but also from sand after a turtle lay on it for 30 min while awaiting treatment procedures (Fig. 2b).
a Correlation of individual patient tumor surface area (mm2) and the concentration of ChHV5 virus being shed into their tank water, as detected by ChHV5 UL30 gene DNA qPCR (positive correlation, Pearson correlation coefficient test, R2 = 0.5431, p = 0.0002, df = 19). Water containing virus schematic insert was generated using BioRender (https://biorender.com/). b Detection of ChHV5 viral shedding onto sand which a patient (“Archie Carr” 49-2020-Cm) temporarily lay on while awaiting treatment. Detection of ChHV5 UL30 gene DNA by qPCR. Amplification ratios for each sand sample are provided in Supplementary Table 1. Error bars denote the standard deviation of six technical replicates. c Patient time-course of tumor surface area changes (surgical removal) and concentration of ChHV5 shed into tank water, as detected by UL30 qPCR. d Time-course of ChHV5 viral shedding into four patient tanks, as detected by UL30 qPCR. Tumor removal surgery events are denoted by an asterisk. Note: the ChHV5 eDNA detected in tank 1 in week 4 was due to a second patient (FP-positive) being added to that tank for a single week, due to the rehabilitation needs of the hospital. Error bars denote the standard deviation of three biological samplings, each with three qPCR technical replicates. e Prolonged patient time-course of tumor surface area changes (surgical removal) and concentration of ChHV5 shed into tank water, as detected by UL30 qPCR. f Correlation of Chelonia mydas (green sea turtle) eDNA shedding and ChHV5 eDNA shedding (positive correlation, Pearson correlation coefficient test, R2 = 0.66, p = 0.00001, df = 19), both C. mydas (16S rRNA gene assay) and ChHV5 (UL30 gene assay) eDNA were detected by qPCR.
Importantly, not only was ChHV5 detectible, it was quantifiable, allowing comparisons between individual tanks and across time (Fig. 2a–f). The level of detectible virus in patient tank water was positively correlated to the tumor burden of the patient(s) housed in that tank (Pearson correlation coefficient test, R2 = 0.5431, p = 0.0002, df = 19) (Fig. 2a). Patients with large well-established tumors shed more virus into tank water than those with small new-growth tumors. Larger tumors shed more ChHV5 by virtue of their cumulative size, not because they contain more lytic ChHV5 per unit area than new growth tumors (see transcriptomics section below). As FP tumors were surgically removed, the level of ChHV5 in patient tanks dropped (Fig. 2c–e), suggesting that the tumors are the primary source of environmental ChHV5 (either through direct tumor shedding, or migration of virus throughout the body and excretion in bodily fluids). The level of ChHV5 in tank water of patients with high tumor burdens was higher than that seen in leeches feeding on FP tumors (Figs. 1c–e, 2a,c–e). Additionally, the quantity of ChHV5 eDNA in tank water was positively correlated with the C. mydas (green turtle) eDNA level (Pearson correlation coefficient test, R2 = 0.66, p = 0.00001, df = 19), as detected by a custom C. mydas 16S rRNA DNA assay (Fig. 2f). ChHV5 eDNA was also detectible in the saltwater fishpond (approximate size 661,000 liters, Supplementary Fig. 1a) which receives the filtered outflow sea water from our patient tanks (Supplementary Table 1), highlighting ChHV5’s potential persistence in aquatic environments.
ChHV5 is latent in FP tumors, including early stage, regrowth, and internal tumors
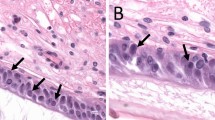
While further investigation of the genomic and environmental drivers of FP, and ChHV5 transmission is warranted31, the suspected causal relationship between ChHV5 and FP also requires further study. Across all our sequenced FP samples (RNA-seq), ChHV5 transcripts were low, and we detected no major switch to active (lytic) virus in either new growth FP or post-surgical regrowth FP (Fig. 3a). In fact, levels of ChHV5 transcripts were only marginally (and not significantly) higher in FP tumors than they were in non-tumor tissue controls (Fig. 3a). The only significant difference found in the level of viral RNA transcripts between any of the groups was between regrowth and established growth external tumors (Kruskal–Wallis with Dunn–Bonferroni post hoc, p = 0.013). Furthermore, we detected no switch to active (lytic) viral transcription in internal tumors (Fig. 3a). Supporting the paucity of lytically replicating virus, no inclusion bodies were detected in either internal or external tumors (Supplementary Table 2) by hematoxylin and eosin staining (Fig. 3b). Together this suggests that the role of lytic ChHV5 in FP is minimal and that if ChHV5 is contributing to FP oncogenesis, it is either (i) occurring transiently, during very early tumorigenesis (before visible tumor appears), or (ii) through the expression of ChHV5 miRNAs (not assessed here), or (iii) that it is the latently expressed ChHV5 transcripts that are driving oncogenesis. We therefore next examined the individual gene level transcripts to determine which ChHV5 genes were transcriptionally active in FP tumors (albeit at relatively low levels). Samples were grouped into four types: non-tumor, external FP (including established, new growth, and regrowth), kidney FP, and lung FP (Supplementary Data 1). Across all four sample types quite a consistent pattern of ChHV5 gene expression emerged (Fig. 3c, Supplementary Fig. 1b), with only 22 of ChHV5’s 104 genes showing levels of expression above 1 transcript per kilobase million (TPM, Supplementary Data 2). Latency-associated genes, such as F-LANA formed part of this 22 gene group, being consistently expressed across all tumor types (Fig. 3c and Supplementary Fig. 1b). Given the paucity of lytic ChHV5, these 22 ChHV5 genes represent the most likely viral drivers able to contribute to FP initiation and ongoing tumor development and growth; therefore they warrant further functional investigation.
a ChHV5 expression across each sample type. Box plot with Tukey whiskers of the number of ChHV5 reads (RNA-seq) per 10 million total reads per sample. Individual sample values within each sample type are shown by the open points. Significant differences in averages between sample types were determined by a Kruskal–Wallis with Dunn–Bonferroni post hoc test and are denoted by an asterisk (*). Per sample type: non-tumor n = 20, established external tumor n = 35, regrowth external tumor n = 12, new growth external tumor n = 9, kidney tumor n = 6, lung tumor n = 7. b Top: Rehabilitating FP-afflicted juvenile green sea turtle. Image credit: Nancy Condron. Bottom: Hematoxylin and eosin stained fibropapillomatosis tumor, external regrowth tumor. c Total number of samples in which reads (RNA-Seq) for each ChHV5 gene were detected. A gene was counted as detected if a sample had TPM-normalized counts >0 for said ChHV5 gene.
Higher ChHV5 viral transcription is not indicative of poor outcome
Almost all samples with ChHV5 viral reads above 200 per 10 million total reads originated from just three patients (Supplementary Fig. 2a). Interestingly all three of these higher viral transcript patients were successfully rehabilitated and released. We, therefore, investigated whether there was any relationship between number of viral transcripts and rehabilitation outcome. Counter-intuitively, patients with positive outcome (survived and released) on average had samples with statistically significantly higher ChHV5 transcripts (Mann–Whitney U Test, p = 0.03), while those patients that died in care or were euthanized due to advanced disease actually had lower ChHV5 transcripts (Fig. 4a). Even when internal tissue samples were removed from the analysis (as all internal samples originated only from the deceased/euthanized category), there remained a significant viral expression difference between the two outcome groups (Mann–Whitney U Test, p = 0.0143).
a Box plot with Tukey whiskers of the number of ChHV5 reads (RNA-Seq) per 10 million total reads between patients based on outcome (released vs died in care/humanely euthanized). Individual sample values within each sample type are shown by the open points. Significant difference in averages between the two outcomes was determined by a Mann–Whitney U test and is denoted by an asterisk (*). N = 69 samples. Per outcome: released = 7 turtles; died/euthanized = 5 turtles. b Box plot with Tukey whiskers of the expression levels of the ChHV5 F-LANA genes (both copies of the F-LANA gene: F-LANA and F-LANA’) in transcripts per million (TPM), by patient outcome and tumor location, as detected by RNA-seq. Individual sample values within each sample type are shown by the open points. c Principal component analysis (PCA) of the viral transcriptomes (minus 28 genes which had 0 reads across all samples, see methods) of all tumor samples (RNA-seq), by tumor location and patient outcome.
We next assessed whether individual viral genes correlated to patient outcome. Twenty-one genes were significantly differentially expressed between tumors from poor outcome (died or euthanized) and good outcome patients, all of which were upregulated in poor outcome tumors (Table 1). However, on closer examination of the poor outcome tumors, it was predominantly viral expression in internal tumors rather than external tumors that was responsible for this differential expression (Table 1 and Fig. 4b, c). Interestingly, the four ChHV5 genes which are atypical of alphaherpesvirus, F-lec1, F-lec2, F-sial, and F-M0465, were more highly expressed in internal tumors (Table 1 and Supplementary Fig. 2b). These four genes are postulated to play a role in viral immune evasion and viral pathogenesis65. Of all of the differentially expressed ChHV5 genes, both genomic copies of the latency-associated F-LANA gene showed the largest fold increase in expression in internal tumors (Table 1 and Fig. 4b).
High ChHV5 viral loads predominantly arise from latent virus
Given the consistently low level of ChHV5 transcripts in FP tumors, we next used whole-genome sequencing (WGS/DNA-seq) to quantify the viral load of ChHV5 to determine whether the low number of transcripts arises due to a lack of virus, or whether large quantities of virus are present within FP tumors with the majority of these being latent (not undergoing active viral replication and transcription). By not conducting any viral enrichment steps, the resulting read numbers give a more reliable indication of the relative abundance of viral DNA compared with host DNA (all within a single sample/library). Viral DNA sequencing reads in the FP tumors covered a broad range (Fig. 5a). Interestingly, the tank water eDNA had a higher viral load than plasma and non-tumor tissue from FP-afflicted turtles (Fig. 5a and Supplementary Fig. 2c). The lung and kidney tumor samples from the same patient (patient 27-2017-Cm) had dramatically different viral loads (Fig. 5a, Table 2). The six tumors (external and kidney) from patient “Yucca” (49-2019-Cm) had a range of ChHV5 reads per 10 million total reads (RPTM) from 1198 to 3127 RPTM. The new growth FP tumor from the patient “Lilac” (25-2018-Cm) had an intermediate viral load of 1,036 RPTM (Table 2 and Fig. 5a). Other new growth tumors from “Lilac” also showed high viral loads (Fig. 5b). Despite the high level of ChHV5 DNA sequencing reads in the lung tumor (3673 RPTM), this same tumor only had a very low level of ChHV5 RNA sequencing reads (68 RPTM), more closely resembling the read numbers of the non-tumor tissues (Table 2).
a ChHV5 abundance (WGS DNA-based) across each sample type. Graph of the number of ChHV5 reads (DNA-seq) per 10 million total reads of 35 samples, with Tukey whiskers. Per sample type: external fibropapillomatosis tumor n = 7, internal fibropapillomatosis tumor n = 6 (4 kidney FP, 1 bladder FP and 1 lung FP), non-tumor n = 8 (2 skin, 2 kidney, 1 bladder, 1 lung, and 2 whole blood), plasma from FP-afflicted turtles n = 10, tissue from non-tumor hatchlings n = 3 (1 green, 1 loggerhead, and 1 leatherback), tank environmental DNA (eDNA) n = 1 (pooled sample of eDNA from 4 tanks with FP-afflicted juvenile green patients and 1 tank with FP-free loggerhead hatchlings). P-values for all groups with significant differences (t-test) to the non-tumor (skin and whole blood) sample cohort are shown on the graph. All samples are from green sea turtles, with the exception of two of the hatchling samples (loggerhead and leatherback), and the eDNA tank sample being pooled from four green and one loggerhead tank water extractions (Supplementary Fig. 2c). b Image of new growth tumors on patient “Lilac” (25-2018-Cm). “Lilac” was admitted to the hospital without tumors, but with a number of leech bites which later developed into FP tumors. ChHV5 relative quantification of “Lilac” new growth tumor samples and non-tumor skin punch biopsy samples using the UL30 DNA qPCR assay37 (Supplementary Table 6). Error bars denote the standard deviation of three qPCR technical replicates. ChHV5 DNA was detected in these growths via qPCR prior to them being classified as fibropapillomatosis by histology. c ChHV5 viral quantification box plot with Tukey whiskers of a range of FP types (n = 43 samples) and non-tumor tissue (n = 36) samples from 13 individual patients using the UL30 DNA qPCR assay37 (Supplementary Table 6). Individual sample values within each sample type are shown by the open points. Absolute quantity of ChHV5 was determined through a standard curve of known amounts (in picograms) of a UL30 gene fragment (Supplementary Table 6). Truncated x-axis label, internal sub-cutane. = internal sub-cutaneous.
The kidney tumor from patient 27-2017-Cm had minimal viral load (Table 2 and Fig. 5a) and this same tumor had minimal copy number variations in the host tumor genome30, suggesting that the tumor must be driven by other oncogenic mechanisms, such as point mutations, epigenetic changes, or transcriptional/translational aberrations. The viral load of this kidney tumor was within the range of non-tumor tissues (juveniles and hatchlings) and plasma from FP-afflicted turtles (Fig. 5a).
When DNA-based viral reads (Fig. 5a) were compared with viral reads from the RNA transcripts (Fig. 3a) of the 90 RNA-seq samples, the highest viral transcript load observed was ~600 RPTM (one FP and one non-tumor sample), while the highest viral DNA load observed was 3,673 RPTM. Interestingly, a high viral DNA load did not equate to high viral transcription (RNA) in the same sample (Table 2). As expected, WGS reads were dispersed across the entire ChHV5 genome (Supplementary Fig. 3a), not being restricted to ChHV5’s transcriptionally active regions (Supplementary Fig. 1b). This confirms that the limited transcriptional signature is not due to a sequencing artifact. Together, the ChHV5 genome-level and gene-level TPM analysis highlight the marked difference in reads between viral DNA presence and viral RNA transcription in FP tumors. Conversely, the range of viral DNA and viral RNA within non-tumor tissue was largely overlapping suggesting that the ChHV5 present in non-tumor samples may be more likely to be transcriptionally active (Table 2 and Figs. 3a, 5a).
Lung and urinary bladder tumors tend to have high viral loads
We next examined the ChHV5 viral loads (viral DNA-based qPCR, ChHV5 UL30 assay) in a wider cohort of internal tumor and matched non-tumor tissue types. Liver and kidney FP tumors had consistently lower viral loads than was seen in other tumor types, although wide ranges in ChHV5 loads were observed in other tumors (Fig. 5c), and some sequenced kidney tumors (all three from patient “Yucca”) had high viral loads (Fig. 5a). Lung and urinary bladder FP tumors consistently had the highest viral loads (Fig. 5c), while, of the non-tumor tissue types assayed, bladder tissue also showed high viral loads, with detected levels being higher than in some of the FP tumors (Fig. 5c).
ChHV5 may be vertically transmitted from mother to offspring
Interestingly, the range of ChHV5 detected in hatchlings (one C. mydas, one loggerhead [Caretta caretta] and one leatherback [Dermochelys coriacea]) overlapped the range of ChHV5 in FP-afflicted non-tumor tissue, the FP kidney tumor, and FP-afflicted blood plasma samples (Fig. 5a). Of the three species, the leatherback sample had the highest number of ChHV5 reads, with many reads aligned to non-coding regions of the ChHV5 genome (Fig. 5a and Supplementary Fig. 4a). ChHV5 DNA was also detected at low levels in six samples from a number of tissues (eye lid, neck, cloaca, front flipper, and heart) from a deceased unhatched green sea turtle (Supplementary Fig. 4b), using the ChHV5 UL30 qPCR assay (viral DNA range: 0–2.24E−05 pg/μl. Amplification ratio: 0.125). No ChHV5 DNA was detected from samples of brain, yolk, and intestine from this turtle.
WGS reveals no evidence of papillomavirus (PV1) association with FP tumors
We used our unbiased WGS to assess whether green turtle or loggerhead specific papillomaviruses (CmPV1 and CcPV1) were present in any of our samples, as although present in sea turtles CmPV1 and CcPV1 have not been strongly associated with FP tumors66,67,68. No CmPV1 reads were detected in any tumor samples, skin samples or plasma samples of FP-afflicted turtles (Supplementary Table 3). Furthermore, unlike ChHV5 (Fig. 3a), no shed CmPV1 or CcPV1 was detected in tank water of pooled FP-afflicted juvenile green and FP non-afflicted loggerhead hatchlings (Supplementary Table 3). From all 38 sequenced samples only two reads were detected for CcPV1, both in C. mydas kidney FP tumors from different individuals (Supplementary Table 3). This might be an artifact as the mate pair of these reads (paired-end sequencing), did not align to CcPV1. These two kidney tumors had 75 and 3,127 ChHV5 RPTM respectively (Table 2 and Supplementary Data 1), but only 0.037 and 0.014 CcPV1 RPTM respectively (Supplementary Table 3). Therefore, our WGS provides no evidence for a correlation between the presence of CmPV1 or CcPV1 and FP tumors, from green turtles from Floridian waters.
Discussion
Anthropogenic activities are fueling the acceleration of the sixth mass extinction event69, and human-wildlife conflict and intensive farming pressure are facilitating zoonotic disease pandemics (such as Ebola, SARS, and COVID-19) with the transmission of animal viruses to humans29,70,71,72,73,74. While the rapid environmental changes induced by human activities are increasing viral transmission from animals to humans, they are also altering the consequences of infections in animal populations, which are more susceptible to disease in the absence of healthy habitats and natural population sizes and ranges with their associated robust genetic variation29. Anthropogenic activities are likely increasing cancer rates in wild populations75,76, with increased incidence and range of sea turtle fibropapillomatosis appearing to be a direct result of human activities impairing the ability of inshore juvenile sea turtles to combat the ChHV5 virus3,31,77. However, studies establishing functional links between environmental contaminants and fibropapillomatosis tumorigenesis, especially across large spatial scales, are required.
It is thought that environmental changes are key to conferring oncogenicity upon ChHV5, potentially through compromising the immune system of sea turtles3,10,78,79. While hosts attempt to mount an immune response30 tumors still develop in a large proportion of individuals3,5,9. The causal link between ChHV5 and fibropapillomatosis also requires further scrutiny. As revealed here, either the role of ChHV5 in driving FP tumor formation and progression is restricted to extremely early tumorigenesis events, or it is driven by latently expressed genes, or is overstated.
As a number of ChHV5 strains exist, it was initially hypothesized that some strains were more oncogenic than others, thus accounting for differences of the severity of FP disease between individuals and populations. However, it has consistently been shown that different ChHV5 strains bear no correlation to disease severity3,41,44,46,80, although large scale geographic differences do exist.
Similar to smaller cohorts of external FP tumors assessed by RNA-seq5,81, we found no evidence to support widespread lytic ChHV5 viral replication within any of the tumor types profiled here. This includes early stage new growth tumors, suggesting that the viral ‘hit and run’ hypothesis82 of oncogenic transformation does not apply to fibropapillomatosis tumors, or that if it does it must be extremely temporally restricted, likely before a visible tumor develops. Furthermore, our results reveal that if ChHV5 is responsible for inducing de novo tumor formation in internal organs, then the limited expression and load of ChHV5 in some internal tumors is highly irregular, particularly in kidney and liver tumors. That even tumor samples with high ChHV5 loads (DNA-level) have only low ChHV5 gene expression (RNA-level), suggests that rapid proliferation of host tumor cells infected with latent virus is the primary driver of the high viral loads sometimes observed in FP tumors. Fibropapillomatosis cell proliferation can be rapid and tumors can double in size in less than two weeks1. Similarly, the paucity of elevated levels of viral transcripts across all tumor samples, in addition to the lack of inclusion bodies, suggests that lytic virus may not be driving the establishment of numerous primary tumors within the same individual. The presence of inclusion bodies in FP tumors is relatively rare. In one study, from the analysis of 381 FP tumors from 17 individuals, epidermal intranuclear inclusion bodies were only identified in 35% of FP-afflicted individuals, and even within that subset of individuals, only 7% of their tumors contained at least one inclusion body48. Similarly, in situ hybridization of the ChHV5 UL30 gene only detected expressed ChHV5 transcripts in three out of 25 tumors83. Even within those three tumors, there was only a low level of expression which was spatially restricted83. Processes occurring in the dermis (especially relating to fibroblasts) may also influence the development of tumors and host-viral interactions, as FP tumors involve changes to both epidermal and dermal cells. Our whole-genome viral transcriptomics confirms that lytic ChHV5 gene expression is lacking in both external and internal tumors. Even while in their latent stage oncogenic viruses can manipulate host cell signaling and contribute to oncogenic transformation and tumor development84,85,86.
Counterintuitively, we show that animals with lower overall viral transcriptional expression had worse rehabilitation outcomes than those with higher viral transcription. Oncogenic viruses can use latency as an immune avoidance strategy, with bouts of temporally and spatially restricted lytic activity helping to prevent a largescale immune response. This feature of oncogenic viruses may help to explain why ChHV5 is predominantly latent in FP tumors, and why lytic viral replication is so spatially restricted48,83. In this context, FP tumors with higher ChHV5 expression levels may be more prone to inducing immune responses, leading to improved patient outcomes. Further supporting this hypothesis, we recently showed that, in the same patient cohort used in this study, higher expression levels of immune-related host genes were associated with better patient outcomes38.
The latency-associated gene LANA and the four atypical ChHV5 genes (F-lec1, F-lec2, F-sial, and F-M04)65 were more strongly expressed in internal tumors than external tumors, although overall expression levels were relatively low. The differing ChHV5 gene expression profiles between internal and external FP tumors, suggests that the mechanisms driving internal tumor development are different from those driving external tumor development. Supporting this, we recently showed that the host oncogenic signaling events driving internal and external tumor development also differ dramatically38.
Sea turtle papillomaviruses (CmPV1 and CcPV1) were originally described in non-FP proliferative skin lesions66,67. However, a subsequent report described papillomavirus in cell lines derived from FP tumors and postulated that they may be associated with sea turtle fibropapillomatosis tumors68. Fibropapilloma tumors are associated with papillomaviruses in other domesticated and wild species87,88,89, as well as a variety of human cancers, including cervical, and head and neck cancer90,91. However, it remains to be resolved whether papillomaviruses have any correlation to FP tumor occurrence. Using qPCR, CmPV1 was recently detected in skin tumor samples (<10 tumors) of Australian C. mydas, while cloacal swabs, blood, and normal skin samples from the same animals tested negative68. However, when we assessed for the presence of PV1 by WGS we detected no CmPV1 in tumor samples, skin samples, blood plasma or tank water. While a greater number of individuals and locations should be assessed, we found no evidence for PV1 as a causative or even correlative factor to sea turtle fibropapillomatosis, despite the prominent role of papilloma viruses in papilloma tumors of other species87,88,89,90,91. Given their small genome and ability to induce oncogenesis at lower loads than herpesviruses, targeted qPCR approaches should also be employed, to confirm whether papillomaviruses are absent from FP tumors68,92. Recently, such approaches detected PV1 in 47% of FP tumors in Australia93.
ChHV5 DNA was identified in C. mydas, loggerhead (Carretta carretta), and leatherback (Dermochelys coriacea) hatchlings, and their abundance overlapped the range (no significant difference) seen in non-tumor tissue and blood plasma from FP-afflicted turtles, and an FP kidney tumor. Although it should be noted that ChHV5 genome coverage was not as complete in these hatchling samples as for tumor samples (Supplementary Fig. 4a). ChHV5 DNA was also detected at low levels, by qPCR, in a pre-hatchling that died prior to successfully hatching. A key objective should be to confirm whether vertical transmission of the virus occurs from mother to offspring. This hypothesis conflicts with the predominant hypothesis that ChHV5 infection is acquired only after juvenile turtles recruit to nearshore habitats46,94. If widespread vertical transmission is confirmed, the relative contribution of vertical versus horizontal transmission needs to be determined. If hatchlings are already infected with ChHV5 (vertical transmission), this has serious implications for any potential population-level vaccination-based mitigation strategies. On-beach (immediate nest emergence95) sampling and qPCR and sequencing-based ChHV5 detection should be conducted to confirm this finding and to determine the prevalence of ChHV5 infected hatchlings. Non-invasive sampling of sand from nest chambers may also reveal whether ChHV5 is present, given that we could detect ChHV5 from sand samples exposed to FP-afflicted patients.
In terms of horizontal vector-borne transmission, we show that ChHV5 was detectible in 90% of leeches removed from FP tumors, this contrasts to a reported 27.5% of leeches from FP-afflicted turtles testing positive for ChHV596. This difference likely arises from relatively rapid testing (3 days to 7 months) in a rehabilitation setting, as opposed to the historical frozen sample set that was used for the Rittenburg et al. 2021 study96. Leeches likely only represent a mechanical vector of ChHV5 transmission, as studies to determine whether leeches function as a true vector with ChHV5 replicating within leeches (as an intermediary host) have not yet been conducted. While a broad range of ChHV5 quantity was detected across leech and patient tank water samples, ChHV5 tended to be more abundant in the water column than in marine leeches. This suggests that both direct water-borne transmission, and vector-borne transmission of ChHV5 are likely.
Accurate detection and monitoring of wildlife pathogens (both vertical and horizontal transmission) with the capacity to impact species survival is essential to devise and implement appropriate mitigation policies63. Environmental DNA approaches have been shown to detect aquatic pathogens earlier than traditional methods and provide advanced warning of infection and mass mortality events57,63. Our eDNA-based detection of ChHV5 in sea water is particularly significant given that, unlike more immediate acting pathogens, for virally-induced cancers there is generally a long lag time (years or decades) between infection and tumor formation97,98,99. It has previously been asserted that only a small percentage of FP tumors shed virus (7% of tumors in 35% of individuals)48. However, our eDNA-based monitoring of viral shedding demonstrated that ChHV5 could be detected even in the tanks of turtles with low tumor burdens and that ChHV5 shedding positively correlated to tumor burden. Other disease-associated green turtle herpesviruses have been shown to remain infective after exposure to sea water, though this has not yet been assessed for ChHV5100. Our findings regarding the predominance of latent ChHV5 within fibropapillomatosis would suggest that not all viral DNA in patient tank water may derive from lytically produced virus. Future studies should determine what proportion of detectible ChHV5 eDNA in patient tanks is derived from viral DNA within host cells compared with free viable infective virus. That urinary bladder samples had high viral loads has potential implications for the spread of ChHV5, as urine is another potential source of ChHV5 release from the body and transmission49,50.
In the near future, eDNA technologies57,63 may allow for the early detection of ChHV5 presence in the environment of vulnerable populations (akin to our detection of ChHV5 in the larger fishpond), and enable further research into the etiology, host species transmission and disease ecology of FP. This unique tool to dynamically track environmental viral shedding in response to disease progression and clinical interventions such as surgery and drug treatment will greatly enhance our ability to address fundamental questions relating to this disease, as well as to design evidence-based rational management strategies (e.g., containment/isolation policies). The approach can also be used for regular or intermittent monitoring of tank water in captive turtle populations, for early warning and disease prevention strategies. Such approaches can also improve our understanding of how environmental and clinical factors influence viral release and spread. The ability to perform eDNA-based quantitative viral shedding monitoring will also further enhance the utility of FP as a model not just to better understand this wildlife epizootic, but also to address unresolved questions relating to viral shedding for other animal and human pathogen-induced cancers.
Summary
Taken together, our results provide transcriptome and genome-level profiling of ChHV5 across external, new growth, established, post-surgical regrowth, and internal visceral tumors (Table 3). They reveal the paucity of lytic ChHV5 replication within tumors, even those harboring high viral loads. High viral loads, where they do occur, likely represent latent virus being produced as infected tumor cells rapidly proliferate. They also revealed that a number of tumors, particularly some internal types, have extremely limited viral loads (DNA-based assessment), calling into question the role of ChHV5 in the establishment and growth of these tumors. The application of novel eDNA-based pathogen monitoring revealed that ChHV5 is shed into the water column, and quantification of ChHV5 eDNA revealed that the level of virus in patient tank water correlates to patient tumor burden. ChHV5 shedding is prevented upon excision of fibropapillomatosis tumors. Improved understanding of the oncogenic role of ChHV5 and its transmission can inform population management and clinical management strategies, including informing biosecurity and quarantine practices.
Methods
Tissue sampling
Sampling was carried out under permit number MTP-21-236 from the Florida Fish and Wildlife Conservation Commission and with ethical approval from the University of Florida’s Institutional Animal Care and Use Committee (IACUC). External fibropapillomatosis (FP) tumors were surgically removed by laser resection as part of the turtle’s rehabilitative care or obtained during necropsy5. Non-tumored areas of the same turtles were obtained by 4 mm punch biopsies during the tumor removal surgery, or during necropsy. Non-tumored sites were selected by gross examination of the region by the attending veterinarian and confirmed visually to be tumor-free normal skin regions and not bordering any tumorous regions by the attending vet, technicians, and researchers. External tumors were sampled from a range of body locations, including skin, plastron, and eye tumors (Supplementary Data 1). Internal tissue samples (tumors and non-tumor tissue samples from lung, kidney, bladder, liver, mouth, sub-cutaneous, and heart) were obtained from animals during necropsies conducted immediately after euthanasia. Note that no animal was euthanized for the purposes of this study, but rehabilitating sea turtles found to harbor internal tumors are currently euthanized in Florida, according to Florida Fish and Wildlife Conservation Commission protocols, as no treatment yet exists for internal tumors. Additional complications arising from surgery and other health concerns may also sometimes necessitate the humane euthanization of sea turtles in rehabilitation. Internal tissue samples were treated the same as the external samples. All samples were obtained from juvenile C. mydas, as this life stage is the most commonly afflicted by the disease. Sex is not readily determinable in juveniles, but was provided for individuals that were euthanized due to internal tumors or other complications and in which necropsies were performed, or for individuals that were endoscoped (KARL STORZ, Multi-Purpose Rigid Endoscope for small animals) as part of their rehabilitative care (see Supplementary Data 1). Samples were stored in RNA-later (Qiagen) at −80 °C, according to the manufacturer’s instructions, until extraction. Samples were stored between <1 day and 8 months, with 85% of samples being extracted within one month.
For the three hatchling samples (one green, one leatherback, and one loggerhead) utilized for WGS, these were admitted to our rehabilitation hospital but died in care. They were housed completely separately to FP-afflicted juvenile patients and frozen at −20 °C upon death. Within two months the carcasses were thawed and during necropsy a cross section of the front flipper of each hatchling was used for DNA extraction (see below). This sampling was conducted using sterile technique, with separate instruments (single use disposable), and was conducted in a lab which never housed FP-patients or any patients from other size classes. For the unhatched green sea turtle utilized for qPCR, upon death this animal was necropsied (on the same day) and tissue samples were obtained from the following locations; eye lid, neck, cloaca, front flipper, heart, brain, yolk, and intestine, and DNA extracted immediately (see below). Prior to death, this turtle remained in its shell and never encountered FP-afflicted turtles, areas, or instruments.
Leeches were removed from six patients during routine admission exams and stored in RNAlater (Qiagen) at −80 °C until extraction. Leeches used for pooled leech extractions were stored for up to 7 months prior to extraction (from “Ruth Gates”, 07-2015-Cm and 09-2015-Cm were stored for 1 month, from “Bruno Hofer” for 5 months, and from “Richard Dawkins” for 7 month). Leeches used for individual leech extractions (from “Broccoli”) were stored for 3 days prior to extraction. Leeches were removed from FP tumors, from non-tumor sites (soft skin/plastron) of FP-afflicted turtles, or from non-FP afflicted turtles. Each entire leech (individual samples) or groups of approximately 10 entire leeches (pooled samples, from the same location type) including the blood pellet were homogenized and used for DNA-extraction (using a DNeasy Blood and Tissue Kit [Qiagen, Cat No. 69504], see below). The majority of the leeches were removed from inguinal regions (both tumor and non-tumor), all 30 leeches extracted individually were removed from two FP tumors on patient “Broccoli’s” ventral inguinal region (Fig. 1a).
RNA and DNA Isolation, library preparation, and sequencing from tissue and eDNA samples
All sequencing was conducted at the University of Florida’s Interdisciplinary Center for Biotechnology Research Core Facilities. For RNA-Seq samples, total RNA was extracted using either an RNeasy Fibrous Tissue kit (Qiagen, Cat No. 74704) or RNeasy Plus kit (Qiagen, Cat No. 74134) with column-based genomic DNA removal, according to the manufacturer’s instructions. Ninety RNA samples, comprising 70 FP tumor samples and 20 non-tumor samples from 12 juvenile green turtles which had stranded in Northern Florida, were used for sequencing. Samples were further categorized by tissue type, as well as growth profile for the external tumors only (see Supplementary Data 1). Sequencing libraries were generated from 500 ng of total RNA using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Cat No. E7530), including polyA selection, according to manufacturer’s protocol. Size and purity of the libraries were analyzed on a Bioanalyzer High Sensitivity DNA chip (Agilent). The RNA samples used for library construction had a RIN value range of 7.2 to 9.8, with the median RIN value of all samples being 9.1. Libraries were sequenced as paired-end reads with a read length of 100 bp on a HiSeq 3000 (Illumina). ERCC Spike-In Mix (ThermoFisher) was used as an internal control: 2 μL of 1:400 diluted ERCC Spike-In Mix with 500 ng of total RNA input.
Fibropapillomatosis-afflicted juvenile C. mydas tissue DNA-seq samples were sequenced in two batches, 6 initial samples on a HiSeq 3000, and 18 NovaSeq 6000 (Illumina) samples. For HiSeq 3000 samples, DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen, Cat No. 69504). Libraries were generated using Illumina TruSeq DNA PCR-Free Library Prep kit including fragmentation with a Covaris S220 sonic disruptor. Size and purity of the libraries were analyzed on a Bioanalyzer High Sensitivity DNA chip (Agilent). Libraries were sequenced as paired-end reads with a read length of 100 bp on an Illumina HiSeq 3000. Six whole genomic DNA samples, comprising three FP tumor samples and patient-matched non-tumor samples from two juvenile green turtles which had also stranded in Northern Florida, were used for sequencing. These samples were further categorized by tissue type, with one tumor and patient-matched healthy tissue sample each coming from an external, kidney, and lung tissue source (see Supplementary Data 1). For NovaSeq 6000 samples (Supplementary Data 1), the 18 DNA samples were fragmented on a Covaris S220 sonicator, to generate ~300 bp fragments. Fragmentation was followed by AMPure clean-up (0.9:1.0 beads:sample ratio). The cleaned-fragmented material (50 ng) was used as input for library construction. Sequencing libraries were constructed using the NEBNext UltraTMII DNA Library Prep Kit for Illumina (Cat# E7645S), according to the manufacturer’s protocol. A set amount of 10–20 ng of sample was used to barcode and generate full adapter-ligated libraries through PCR. PCR was done for only 6–7 cycles of amplification in order to minimize duplicate reads. Barcoding was done using the indexing reagents provided in the NEBNext Unique Dual Index Oligos kit (Cat# E6440S). The final libraries were quantified by fluorescence (QUBIT, ThermoFisher), and sized on the Agilent TapeStation (DNA5000 Screen Tape). A yield of 50–60 ng of library was obtained of an average size ranging from 460 to 600 bp. Libraries were normalized and pooled equimolarly. This step was followed by treatment with the “Free Adapter Blocking Reagent” protocol (FAB, Cat# 20024145) in order to minimize the presence of adaptor-dimers and index hopping rates. The library pool was diluted to 2.5 nM and sequenced (one S4 lane, 2 × 150 cycles [paired-end]) according to Illumina NovaSeq6000 sequencing protocol, using 180 pM loading concentration and 1% PhiX spike-in control. Approximately 10 billion paired-end reads were obtained for the entire run (Q30% > = 80%; Cluster PF = 70%), ~8.7 billion paired-end reads were generated (NovaSeq 6000) for the 18 C. mydas samples, with the remainder of the reads relating to samples from other projects.
In addition, whole genomic DNA was extracted from 10 plasma samples taken from six individual turtles during the course of their rehabilitation (Supplementary Data 1). Excess blood was utilized from blood samples drawn for routine medical care by our veterinarians and plasma was separated from the red blood cells by centrifugation. Only 100 µl of plasma was collected and stored as allowed by permit number MTP-21-236 from the Florida Fish and Wildlife Conservation Commission. DNA was extracted from 60 µl of plasma using a DNeasy Blood & Tissue Kit (Qiagen, Cat No. 69504) and used to produce low input Illumina Fragment libraries using a Low Input Library Prep kit v2 (Clontech Laboratories, Inc., Catalog No. 634899). DNA was fragmented using the Covaris S220 sonic disruptor and libraries were then sequenced as paired-end reads with a read length of 100 bp on an Illumina HiSeq 3000.
Furthermore, three whole genomic DNA samples were also taken from ground flipper tissue samples from three deceased hatchling turtles (never in contact with FP-afflicted patients or water/tanks) of each separate species: a green (C. mydas), loggerhead (C. caretta), and leatherback turtle (D. coriacea), and processed following the methods for tissue (HiSeq 3000) as detailed above. The leatherback sequence length was the only sample that differed, with paired-end reads of length 150 bp instead of 100 bp. TPM generation of hatchling samples was done using viral aligning reads only, as not all of the turtle species have published reference genomes.
Finally, one pooled library of environmental DNA (eDNA), combining five holding tank water samples from the Whitney Laboratory Sea Turtle Hospital facility of the University of Florida was also used for sequencing. For this pooled sample, water collection, filtration, and extraction were performed separately on each tank sample, and the final purified DNA was pooled prior to library preparation. Pooling was conducted to obtain the average ChHV5 load across the five tanks. Seawater from five tanks (four housing juvenile green sea turtles and one housing loggerhead post-hatchling washbacks, 500 ml seawater per tank) was filtered (EMD Millipore PES 0.22 μm Sterivex filter) by a hand pump and DNA was extracted from the filter using a Qiagen DNeasy Blood & Tissue Kit (Qiagen, Cat No. 69504) and modified extraction protocol was carried out58, with an overnight (24–36 h) incubation of the filter at 56 °C in Buffer ATL and Proteinase K in a hybridization oven with agitation. Libraries were generated using a NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, Cat No. E7645), including fragmentation with a Covaris S220 sonic disruptor. Fragment size and purity of the libraries were analyzed on a Bioanalyzer High Sensitivity DNA chip (Agilent). Libraries were sequenced as paired-end reads with a read length of 100 bp on an Illumina HiSeq 3000.
Quality control and read trimming
The software FastQC—https://www.bioinformatics.babraham.ac.uk/projects/fastqc/—was used to assess data quality. Reads were then trimmed with trim_galore (The Babraham Institute, version 0.5.0) to remove ends with a Phred quality score less than 30, to remove adaptor sequences, and to remove sequences fewer than 25 bp after trimming. For any samples that contained overrepresented sequences according to FastQC, the trimmomatic tool101 (version 0.36) was then used to remove these sequences from reads and any sequences less than 25 bp after trimming. The number of raw reads per sample and reads remaining after trimming can be found in Supplementary Data 1.
Read alignment and read counts
Reads from all samples (RNA-seq, DNA-seq, and eDNA) were first aligned to the ChHV5 genome [GenBank accession number: HQ878327.2]65 to determine the level of ChHV5RNA and DNA present in each sample using bowtie2102 (version 2.3.5.1). The overall alignment rate to the ChHV5 genome was low for both RNA-seq and DNA-seq samples, with most reads aligning to the green turtle genome (NCBI GenBank Accession numbers: GCA_000344595.1 and GCA_015237465.1), as expected (Supplementary Data 1).
Transcript abundance for ChHV5 virus-specific transcripts was generated using htseq-count103 (version 0.6.1p1) with the following parameters: not strand-specific, feature type ‘gene’, intersection non-empty mode, and a minimum aQual of 0. Count tables for viral transcripts were merged for all RNA-seq samples and counts were normalized for gene length and sequencing depth by transcripts per million (TPM) (Supplementary Data 2). TPMs were generated using the combined host (C. mydas30) and ChHV5 reads to allow direct comparisons between host and viral gene expression levels.
Differential expression analysis
Prior to differential expression analysis, a 1 was added to all gene counts to avoid zero counts in the analysis. The following viral genes that had the same counts across all samples were removed due to having a variance of 0 across samples: UL15A, UL15B, UL16, UL20, UL26.5, UL31, UL37, UL53, HP1/HP1′, HP2/HP2′, HP7, HP8, HP11, HP12, HP14, HP15, HP18, HP21, HP22, HP23, HP25, HP26, HP27, HP29, HP31, HP38. An external FP tumor sample, emLFF3H, was also removed from the analysis due to it being an outlier. A principal component analysis (PCA) plot (see Fig. 4c) was generated using the PtR script in the Trinity toolkit104. Differential expression analysis was carried out in the R statistical environment (v 3.4.4) using the quasi-likelihood algorithm within the software package ‘edgeR’ (v 3.20.9)105,106,107. Samples were first grouped by rehabilitation outcome: good outcome (released, n = 46 samples) versus poor outcome (died in care or humanely euthanized, n = 23 samples). The trimmed mean of M-values normalization method was then used to calculate effective library size. To model read counts for each gene, the quasi-likelihood extension for the negative binomial distribution107 was used. Differential expression analysis was then conducted in which good outcome was set as the “treatment” and poor outcome as the “reference” or “control” group. A false discovery rate (FDR) of 5% was used as a cut-off to determine significantly differentially expressed viral genes between rehabilitation outcomes. Results of this analysis are included in Table 1. Boxplots were generated using BoxPlotR108.
Tissue and eDNA qPCR
qPCR assays were conducted on non-tumor tissue and FP tissue samples to quantify viral load within a range of tumor types, as well as water and sand eDNA samples to look at viral shedding dynamics within rehabilitation tanks. DNA was extracted from 79 tissue samples from 14 juvenile green turtles which had stranded in Northern Florida (Supplementary Data 3), 25 tissue samples from an unhatched green sea turtle and from leeches removed from six turtles as detailed above. Leeches from the same location were either pooled prior to extraction (~10 leeches per pool, Fig. 1d), or extracted individually (Fig. 1e, Supplementary Table 1). DNA was also extracted from 50 tank water samples (19 samplings with two-three sampling replicates per sampling event) and an additional 26 tank water samples from one prolonged-duration individual (13 samplings with 2 sampling replicates per sampling event). DNA was also extracted from 1 pond water sample (three sampling replicates), on the surface of the east end of the pond, close to the outflow pipe (Supplementary Fig. 1a). The fishpond is ~661,000 liters. Environmental DNA was extracted as per the eDNA sequencing samples above, with 500 ml seawater filtered per tank, except for the samples in Fig. 4e in which 1-liter seawater per sample was collected. Filtering (EMD Millipore PES 0.22 μm Sterivex filter) was performed by hand pump, except for samples depicted in Fig. 4e for which a Geotech Peristaltic Pump was used.
For sand eDNA samples, a 50 ml Falcon conical centrifuge tube was filled with sand from the base of a plastic holding box used to house an FP-afflicted juvenile Chelonia mydas (“Archie Carr” 49-2020-Cm) (on two separate occasions, approximately one month apart). The patient was housed in the box for 30 min, (dry-docked) during routine rehabilitative treatment at the Whitney Sea Turtle Hospital. While awaiting examination or receiving treatment, patients are routinely dry-docked for short periods of time. The 50 ml sand samples were sub-divided into 2–3 10 ml biological replicates. Each replicate was then washed in 20 ml 1× IDTE pH 8.0 TE Solution in individual 50 ml Falcon conical centrifuge tubes. Samples were shaken gently by hand then set on a rocking platform for 1 h at room temperature, shaking gently by hand every 15 min. Samples were then rested until sand sunk to the bottom of each tube (~30 s), then the aqueous layer was immediately pipetted into a 60 ml sterile BD luer lock syringe. Samples were hand filtered through 0.22 µm Sterivex-GP Pressure Filter Units and capped with B.Braun luer lock caps. 740 µl Buffer ATL and 60 µl Proteinase K from a Qiagen DNeasy Blood and Tissue Kit were added to each sample and eDNA was extracted as described for water eDNA samples (see above).
ChHV5 viral load was quantified using TaqMan Fast Advanced Mastermix (ThermoFisher, Cat No. 4444557) according to the manufacturer’s protocol by amplifying the ChHV5 virus-specific DNA polymerase (UL30) gene37 (Supplementary Table 4). A species-specific assay was also developed to target the 16S ribosomal RNA gene (rRNA) for Atlantic populations of C. mydas to serve as positive controls and to compare the level of viral DNA present against the level of C. mydas DNA present within each sample (Supplementary Table 4). This sea turtle species was the primary focus as they are the ones that are most commonly housed at the Whitney Sea Turtle Hospital and most commonly afflicted by FP. A LightCycler480 Instrument II (Roche) was used for amplification and cycling parameters were as follows: 95 °C 10 min, 45 cycles of 95 °C 10 s, 60 °C 20 s, and 72 °C 20 s, or an Applied Biosystems QuantStudio 6 Pro was used for amplification and cycling parameters were as follows: 50 °C for 2 min, 95 °C for 10 min, 45 cycles of 95 °C for 15 s and 60 °C for 1 min. All samples were run in triplicate with negative controls, except sand eDNA samples, fishpond water eDNA samples, leech samples, and unhatched tissue samples which were run with six technical replicates. Both absolute quantification and relative quantification methods were utilized in this study. Standard curves using synthetic fragments of the UL30 gene and C. mydas 16S rRNA gene (see Supplementary Table 4) were generated to calculate the amount of DNA of these two genes present within each sample (in pg of DNA). For relative quantification, the ΔCt method was used, in which a single sample was set as a reference point to which all other samples were compared.
Tumor burden calculations, statistics, and reproducibility
Tumor burdens were assessed by total tumor surface area using ImageJ (https://imagej.nih.gov/ij/)1. ImageJ image analysis was used to make accurate measurements of tumor two-dimensional surface area, from images taken using an Olympus Tough TG-4 a~30 cm from each lesion, with a 25 cm scale bar for accurate pixel comparison.
For comparison of viral loads (Fig. 5a), data that was normally distributed was compared using t-tests, for comparison of data that were not normally distributed (even after log transformation), non-parametric tests (Mann–Whitney U Test) were performed. Significance was considered to be p values ≤ 0.05. For correlation of tumor burden with viral shedding, and viral and green turtle eDNA a Pearson Correlation Coefficient test was used (Fig. 2a,f). The underlying tumor surface area and ChHV5 concentration data used for the correlation reported in Fig. 2f, is present in Supplementary Data 4. For comparison of viral loads among different groups of RNA-seq (Fig. 3a) samples (non-tumor, established FP, regrowth FP, new growth FP, kidney FP, and lung FP), a Kruskal–Wallis test followed by a Dunn–Bonferroni post hoc to indicate significant differences among groups was performed in SPSS (IBM SPSS®).
Histology methodology, embedding, sectioning, and staining
Turtle tissue samples were surgically removed using a CO2 laser and stored in 4% paraformaldehyde at 4 °C overnight. Samples were washed twice in 1× PBS for 10 min, once in Milli-Q H2O for 10 min, twice in 50% ethanol for 15 min, twice in 90% ethanol for 15 min, and twice in 100% ethanol for 15 min. Samples were then stored in 100% ethanol at 4 °C for 5 nights. Samples were washed in 100% aniline for 1 h, 50:50 aniline:methyl salicylate for 1 h, and twice in 100% methyl salicylate for 1.5 h. Samples were then stored in 50:50 methyl salicylate:paraffin at 60 °C overnight. Samples were washed twice in 100% paraffin at 60 °C for 3 h. Samples were then stored in 100% paraffin overnight. Finally, samples were embedded in 100% paraffin and stored at 4 °C. Paraffin blocks were sectioned into 6 μm ribbons of six tumor sections each, on charged Fisherbrand Superfrost Plus microscope slides using an AO Spencer ‘820’ microtome and stored at room temperature.
For hematoxylin and eosin (H&E) staining, sectioned slides underwent a series of washes: two 7-min washes in xylene, two 7-min washes in 100% ethanol, two 7-min washes in 95% ethanol, 2 min in distilled H2O, 2 min in hematoxylin, two 15-s washes in distilled H2O, 2 min in distilled H2O, 3 min in NH3, 2 min in distilled H2O, 2 min in 95% ethanol, 1 min in eosin, two 2-min washes in 95% ethanol, two 2-min washes in 100% ethanol, and finally two 2-min washes in xylene. Fisher Scientific Permount mounting media was added to stained slides, a cover slip was placed on top, and slides were left to dry overnight before imaging on the Zeiss M2 Axio Imager.
Features of tumors were evaluated by a veterinary pathologist for comparison with the molecular analysis (Supplementary Table 2). Sections were examined for intranuclear viral inclusion bodies. The cellularity of the dermal component of cutaneous tumors and internal tumors was subjectively scored, by the pathologist, on a scale of 1 (sparsely cellular) to 3 (densely cellular). The presence of lymphocytic inflammation within the tumors also scored from 0 (absent) to 3 (abundant). The presence of ulceration of the epidermis and associated heterophilic (granulocytic) inflammation also was noted. These features were selected as they may represent aspects of tumor development, regression, or host response relevant to our analysis.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The RNA-Seq and DNA-Seq data including raw reads are deposited in NCBI (https://www.ncbi.nlm.nih.gov/) under BioProject ID: PRJNA449022.
References
Farrell, J., Thomas, R., Martindale, M. Q. & Duffy, D. J. Characterisation of fibropapillomatosis tumour growth profiles in green sea turtles (Chelonia mydas). Testudo 8, 12–29 (2018).
Williams, E. H. Jr. et al. An epizootic of cutaneous fibropapillomas in green turtles Chelonia mydas of the Caribbean: part of a panzootic? J. Aquat. Anim. Health 6, 70–78 (1994).
Jones, K., Ariel, E., Burgess, G. & Read, M. A review of fibropapillomatosis in Green turtles (Chelonia mydas). Vet. J. 212, 48–57 (2016).
Mascarenhas, R. & Iverson, P. J. Fibropapillomatosis in stranded green turtles (Chelonia mydas) in Paraiba State, Northeastern Brazil: evidence of a Brazilian epizootic. Mar. Turt. Newsl. 120, 3–6 (2008).
Duffy, D. J. et al. Sea turtle fibropapilloma tumors share genomic drivers and therapeutic vulnerabilities with human cancers. Commun. Biol. 1, 63 (2018).
Reséndiz, E. et al. Molecular characterization of chelonid alphaherpesvirus 5 in a black turtle (Chelonia mydas) fibropapilloma from Baja California Sur, Mexico. Animals 11, 105 (2021).
Álvarez-Varas, R. et al. First report of fibropapillomatosis in an olive ridley turtle Lepidochelys olivacea from the southeastern Pacific. Dis. Aquat. Org. 135, 43–48 (2019).
Suárez-Domínguez, E. A. et al. Fibropapillomatosis in free-ranging green sea turtles (Chelonia mydas) off the central coast of Veracruz, Mexico. Cienc. Mar. 46, 133–143 (2020).
Hargrove, S., Work, T., Brunson, S., Foley, A. M. & Balazs, G. Proceedings of the 2015 International Summit on Fibropapillomatosis: Global Status, Trends, and Population Impacts. 85 (2016).
Page-Karjian, A. et al. Factors influencing survivorship of rehabilitating green sea turtles (Chelonia mydas) with fibropapillomatosis. J. Zoo. Wildl. Med. 45, 507–519 (2014).
Duarte, A. et al. First histological and virological report of fibropapilloma associated with herpesvirus in Chelonia mydas at Príncipe Island, West Africa. Arch. Virol. 157, 1155–1159 (2012).
Foley, A. M., Singel, K., Hardy, R., Bailey, R. & Schaf, S. Distributions, relative abundances, and mortality factors for sea turtles in Florida from 1980 through 2007 as determined from strandings (Florida Fish and Wildlife Conservation Commission, 2007).
Aguirre, A. A. & Lutz, P. Marine turtles as sentinels of ecosystem health: is fibropapillomatosis an indicator? EcoHealth 1, 275–283 (2004).
International, C. in Invasive Species Compendium: Fibropapillomatosis of Sea Turtles (2019).
Tavares Martins, S., Peteiro, L. & Marco, A. First documented record of fibropapillomas on green turtles Chelonia mydas in the Cabo Verde Archipelago, West Africa. Zoologia Caboverdiana 8, 14–16 (2020).
Loganathan, A. L., Palaniappan, P. & Subbiah, V. K. Evidence of chelonid herpesvirus 5 (ChHV5) in green turtles Chelonia mydas from Sabah, Borneo. bioRxiv, 2021.2001.2025.428031. Preprint at https://doi.org/10.1101/2021.01.25.428031 (2021).
Stacy, B. A. et al. Report of the technical expert workshop: developing recommendations for field response, captive management, and rehabilitation of sea turtles with fibropapillomatosis. https://doi.org/10.25923/adg2-g771 (2019).
Shaver, D. J., Walker, J. S. & Backof, T. F. Fibropapillomatosis prevalence and distribution in green turtles Chelonia mydas in Texas (USA). Dis. Aquat. Org. 136, 175–182 (2019).
Foley, A. M. et al. Distributions, relative abundances, and mortality factors of sea turtles in Florida during 1980–2014 as determined from strandings. (Fish and Wildlife Research Institute, Jacksonville Field Laboratory, Jacksonville, Florida, USA, 2015).
da Silva-Júnior, E. S. et al. Stranded marine turtles in Northeastern Brazil: incidence and spatial–temporal distribution of fibropapillomatosis. Chelonian Conserv. Biol. https://doi.org/10.2744/ccb-1359.1 (2019).
Loureiro, N. D. S. & Matos, D. Presence of fibropapillomatosis in green turtles Chelonia mydas at Príncipe Island in the Gulf of Guinea. Arquipélago-Life Mar. Sci. 26, 79–83 (2009).
Cruz Sr., E. Saga of the Sea Turtle. The Warty Green Turtle, 287–293 (Privately-published 1985).
Foley, A. M., Schroeder, B. A., Redlow, A. E., Fick-Child, K. J. & Teas, W. G. Fibropapillomatosis in stranded green turtles (Chelonia mydas) from the eastern United States (1980–98): trends and associations with environmental factors. J. Wildl. Dis. 41, 29–41 (2005).
Smith, G. C. & Coates, C. W. Fibro-epithelial growths of the skin in large marine turtles Chelonia mydas. Zoologica 23, 93–98 (1938).
Orós, J. & Torrent, A. Unusual tumors in three loggerhead sea turtles (Caretta caretta) stranded in the Canary Islands. Spain Mar. Turt. Newsl. 88, 6–7 (2000).
Orós, J. et al. Multicentric lymphoblastic lymphoma in a loggerhead sea turtle (Caretta caretta). Vet. Pathol. 38, 464–467 (2001).
Orós, J., Tucker, S., Fernández, L. & Jacobson, E. R. Metastatic squamous cell carcinoma in two loggerhead sea turtles Caretta caretta. Dis. Aquat. Org. 58, 245–250 (2004).
Quesada, V. et al. Giant tortoise genomes provide insights into longevity and age-related disease. Nat. Ecol. Evolution 3, 87–95 (2019).
Whilde, J., Martindale, M. Q. & Duffy, D. J. Precision wildlife medicine: applications of the human-centred precision medicine revolution to species conservation. Glob. Change Biol. 23, 1792–1805 (2017).
Yetsko, K. et al. Mutational, transcriptional and viral shedding dynamics of the marine turtle fibropapillomatosis tumor epizootic. bioRxiv, 2020.2002.2004.932632. Preprint at https://doi.org/10.1101/2020.02.04.932632 (2020).
Duffy, D. J. & Martindale, M. Q. Perspectives on the expansion of human precision oncology and genomic approaches to sea turtle fibropapillomatosis. Commun. Biol. 2, 54 (2019).
Schulz, T. F. & Cesarman, E. Kaposi sarcoma-associated herpesvirus: mechanisms of oncogenesis. Curr. Opin. Virol. 14, 116–128 (2015).
Davidson, N. E. et al. AACR Cancer Progress Report 2016. Clin. Cancer Res. 22, S1–S137 (2016).
Alfaro-Núñez, A. et al. Further evidence of Chelonid herpesvirus 5 (ChHV5) latency: high levels of ChHV5 DNA detected in clinically healthy marine turtles. PeerJ 4, e2274 (2016).
Herbst, L. H. Fibropapillomatosis of marine turtles. Annu. Rev. Fish. Dis. 4, 389–425 (1994).
Herbst, L. et al. Experimental transmission of green turtle fibropapillomatosis using cell-free tumor extracts. Dis. Aquat. Org. 22, 1–12 (1995).
Page-Karjian, A. et al. Quantifying chelonid herpesvirus 5 in symptomatic and asymptomatic rehabilitating green sea turtles. Endanger. Species Res. 28, 135–146 (2015).
Yetsko, K. et al. Molecular characterization of a marine turtle tumor epizootic, profiling external, internal and postsurgical regrowth tumors. Commun. Biol. 4, 152 (2021).
Whilde, J. et al. Behaviour of juvenile green turtles (Chelonia mydas) before and after fibropapillomatosis tumour removal. Testudo 9, 22–35 (2019).
Work, T. M., Dagenais, J., Weatherby, T. M., Balazs, G. H. & Ackermann, M. In-vitro replication of Chelonid herpesvirus 5 in organotypic skin cultures from Hawaiian green turtles (Chelonia mydas). J. Virol. 91, e00404-00417 (2017).
Morrison, C. L. et al. Genomic evolution, recombination, and inter-strain diversity of chelonid alphaherpesvirus 5 from Florida and Hawaii green sea turtles with fibropapillomatosis. PeerJ 6, e4386 (2018).
Alfaro-Núñez, A. et al. Global distribution of Chelonid fibropapilloma-associated herpesvirus among clinically healthy sea turtles. BMC Evol. Biol. 14, 1–12 (2014).
Sellera, F. P. et al. Investigation of green turtle (Chelonia mydas) cutaneous fibropapillomatosis recurrence rates following diode laser surgery. J. Exot. Pet. Med. 28, 180–184 (2019).
Domiciano, I. G. et al. Chelonid alphaherpesvirus 5 DNA in fibropapillomatosis-affected Chelonia mydas. EcoHealth https://doi.org/10.1007/s10393-019-01412-8 (2019).
Rossi, S. et al. Monitoring green sea turtles in Brazilian feeding areas: relating body condition index to fibropapillomatosis prevalence. J. Mar. Biol. Assoc. U.K., 1–9, https://doi.org/10.1017/S0025315419000730 (2019).
Jones, K. et al. Molecular evidence for horizontal transmission of chelonid alphaherpesvirus 5 at green turtle (Chelonia mydas) foraging grounds in Queensland, Australia. PLoS ONE 15, e0227268 (2020).
Page-Karjian, A. et al. Molecular assessment of chelonid alphaherpesvirus 5 infection in tumor-free green (Chelonia mydas) and Loggerhead (Caretta caretta) Sea Turtles in North Carolina, USA, 2015–2019. Animals 10, 1964 (2020).
Work, T. M., Dagenais, J., Balazs, G. H., Schettle, N. & Ackermann, M. Dynamics of virus shedding and in situ confirmation of chelonid herpesvirus 5 in Hawaiian green turtles with fibropapillomatosis. Vet. Pathol. 52, 1195–1201 (2015).
Page-Karjian, A. et al. Potential non-cutaneous sites of chelonid herpesvirus 5 persistence and shedding in green sea turtles (Chelonia mydas). J. Aquat. Anim. Health 29, 136–142 (2017).
Chaves, A. et al. Examining the role of transmission of chelonid alphaherpesvirus 5. EcoHealth 14, 530–541 (2017).
Greenblatt, R. J. et al. The Ozobranchus leech is a candidate mechanical vector for the fibropapilloma-associated turtle herpesvirus found latently infecting skin tumors on Hawaiian green turtles (Chelonia mydas). Virology 321, 101–110 (2004).
Adams, C. I. et al. Beyond biodiversity: can environmental DNA (eDNA) cut it as a population genetics tool? Genes 10, 192 (2019).
Boussarie, G. et al. Environmental DNA illuminates the dark diversity of sharks. Sci. Adv. 4, https://doi.org/10.1126/sciadv.aap9661 (2018).
Evans, N. T. & Lamberti, G. A. Freshwater fisheries assessment using environmental DNA: A primer on the method, its potential, and shortcomings as a conservation tool. Fish. Res. 197, 60–66 (2018).
Qu, C. & Stewart, K. A. Evaluating monitoring options for conservation: comparing traditional and environmental DNA tools for a critically endangered mammal. Sci. Nat. 106, 9 (2019).
Roussel, J. M., Paillisson, J. M., Treguier, A. & Petit, E. The downside of eDNA as a survey tool in water bodies. J. Appl. Ecol. 52, 823–826 (2015).
Strand, D. A. et al. Monitoring a Norwegian freshwater crayfish tragedy: eDNA snapshots of invasion, infection and extinction. J. Appl. Ecol. 56, 1661–1673 (2019).
Seymour, M. et al. Acidity promotes degradation of multi-species environmental DNA in lotic mesocosms. Commun. Biol. 1, 4 (2018).
Creer, S. et al. The ecologist’s field guide to sequence‐based identification of biodiversity. Methods Ecol. Evol. 7, 1008–1018 (2016).
Bohmann, K. et al. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 29, 358–367 (2014).
Thomsen, P. F. & Willerslev, E. Environmental DNA—an emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18 (2015).
Erickson, R. A., Merkes, C. M. & Mize, E. L. Sampling designs for landscape‐level eDNA monitoring programs. Integr. Environ. Assess. Manage. 15, 760–771 (2019).
Miaud, C., Arnal, V., Poulain, M., Valentini, A. & Dejean, T. eDNA increases the detectability of ranavirus infection in an alpine amphibian population. Viruses 11, 526 (2019).
Monezi, T. A. et al. Chelonid herpesvirus 5 in secretions and tumor tissues from green turtles (Chelonia mydas) from Southeastern Brazil: a ten-year study. Vet. Microbiol. 186, 150–156 (2016).
Ackermann, M. et al. The genome of chelonid herpesvirus 5 harbors atypical genes. PLoS ONE 7, e46623 (2012).
Manire, C. A. et al. Proliferative dermatitis in a loggerhead turtle, Caretta caretta, and a green turtle, Chelonia mydas, associated with novel papillomaviruses. Vet. Microbiol. 130, 227–237 (2008).
Herbst, L. H. et al. Genomic characterization of two novel reptilian papillomaviruses, Chelonia mydas papillomavirus 1 and Caretta caretta papillomavirus 1. Virology 383, 131–135 (2009).
Mashkour, N., Maclaine, A., Burgess, G. W. & Ariel, E. Discovery of an Australian Chelonia mydas papillomavirus via green turtle primary cell culture and qPCR. J. Virol. Methods 258, 13–23 (2018).
Ceballos, G., Ehrlich, P. R. & Raven, P. H. Vertebrates on the brink as indicators of biological annihilation and the sixth mass extinction. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1922686117 (2020).
Gibb, R. et al. Zoonotic host diversity increases in human-dominated ecosystems. Nature https://doi.org/10.1038/s41586-020-2562-8 (2020).
Wide Fund For Nature International. COVID 19: Urgent call to protect people and nature. (Gland, Switzerland., 2020).
United Nations. UN Environment Programme and International Livestock Research Institute. Preventing the next pandemic—Zoonotic diseases and how to break the chain of transmission. (United Nations Nairobi, Kenya, 2020).
Johnson, C. K. et al. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc. R. Soc. B: Biol. Sci. 287, 20192736 (2020).
Coller, J., Teperman, E. & Lettini, M. An industry infected: Animal agriculture in a post-COVID world. (Jeremy Coller Foundation, the FAIRR Initiative, London, England, 2020).
Baines, C. et al. Linking pollution and cancer in aquatic environments: A review. Environ. Int. 149, 106391 (2021).
Giraudeau, M., Sepp, T., Ujvari, B., Ewald, P. W. & Thomas, F. Human activities might influence oncogenic processes in wild animal populations. Nat. Ecol. Evol. https://doi.org/10.1038/s41559-018-0558-7 (2018).
Van Houtan, K. S., Hargrove, S. K. & Balazs, G. H. Land use, macroalgae, and a tumor-forming disease in marine turtles. PLoS ONE 5, e12900 (2010).
Aguirre, A. A., Balazs, G. H., Spraker, T. R. & Gross, T. S. Adrenal and hematological responses to stress in juvenile green turtles (Chelonia mydas) with and without fibropapillomas. Physiol. Zool. 68, 831–854 (1995).
dos Santos, R. G. et al. Relationship between fibropapillomatosis and environmental quality: a case study with Chelonia mydas off Brazil. Dis. Aquat. Organ 89, 87–95 (2010).
Lawrance, M. F., Mansfield, K. L., Sutton, E. & Savage, A. E. Molecular evolution of fibropapilloma-associated herpesviruses infecting juvenile green and loggerhead sea turtles. Virology 521, 190–197 (2018).
Blackburn, N. B. et al. Transcriptomic profiling of fibropapillomatosis in green sea turtles Chelonia mydas from South Texas. Front. Immunol. 12, 410 (2021).
Niller, H. H., Wolf, H. & Minarovits, J. Viral hit and run-oncogenesis: genetic and epigenetic scenarios. Cancer Lett. 305, 200–217 (2011).
Kang, K. I. et al. Localization of fibropapilloma-associated turtle herpesvirus in green turtles (Chelonia mydas) by in-situ hybridization. J. Comp. Pathol. 139, 218–225 (2008).
Young, L. S. & Murray, P. G. Epstein–Barr virus and oncogenesis: from latent genes to tumours. Oncogene 22, 5108–5121 (2003).
Young, L. S., Yap, L. F. & Murray, P. G. Epstein–Barr virus: more than 50 years old and still providing surprises. Nat. Rev. Cancer 16, 789–802 (2016).
Ye, F., Lei, X. & Gao, S.-J. Mechanisms of Kaposi’s sarcoma-associated herpesvirus latency and reactivation. Adv. Virol. 2011, 193860 (2011).
McAloose, D. & Newton, A. L. Wildlife cancer: a conservation perspective. Nat. Rev. Cancer 9, 517–526 (2009).
Pesavento, P. A., Agnew, D., Keel, M. K. & Woolard, K. D. Cancer in wildlife: patterns of emergence. Nat. Rev. Cancer 18, 646–661 (2018).
Roperto, S. et al. Oral fibropapillomatosis and epidermal hyperplasia of the lip in newborn lambs associated with bovine Deltapapillomavirus. Sci. Rep. 8, 13310 (2018).
Zapatka, M. et al. The landscape of viral associations in human cancers. Nat. Genet. 52, 320–330 (2020).
Lajer, C. B. et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br. J. Cancer 106, 1526–1534 (2012).
Sun, Z. et al. Development of a fluorescence–based multiplex genotyping method for simultaneous determination of human papillomavirus infections and viral loads. BMC Cancer 15, 860 (2015).
Mashkour, N., Jones, K., Wirth, W., Burgess, G. & Ariel, E. The concurrent detection of chelonid alphaherpesvirus 5 and Chelonia mydas papillomavirus 1 in tumoured and non-tumoured green turtles. Animals 11, 697 (2021).
Ene, A. et al. Distribution of chelonid fibropapillomatosis-associated herpesvirus variants in Florida: molecular genetic evidence for infection of turtles following recruitment to neritic developmental habitats. J. Wildl. Dis. 41, 489–497 (2005).
Perrault, J. R. & Coppenrath, C. M. Albinism in Florida green turtle (Chelonia mydas) hatchlings: ratio-based evidence of basic mendelian recessiveness. Mar. Turtle Newsl.156, 38–40 (2019).
Rittenburg, L. T., Kelley, J. R., Mansfield, K. L. & Savage, A. E. Marine leech parasitism of sea turtles varies across host species, seasons, and the tumor disease fibropapillomatosis. Dis. Aquat. Org. 143, 1–12 (2021).
Münz, C. Latency and lytic replication in Epstein–Barr virus-associated oncogenesis. Nat. Rev. Microbiol. 17, 691–700 (2019).
Ljubojevic, S. & Skerlev, M. HPV-associated diseases. Clin. Dermatol. 32, 227–234 (2014).
Sanclemente, G. & Gill, D. Human papillomavirus molecular biology and pathogenesis. J. Eur. Acad. Dermatol. Venereol. 16, 231–240 (2002).
Curry, S. S. et al. Persistent infectivity of a disease-associated herpesvirus in green turtles after exposure to seawater. J. Wildl. Dis. 36, 792–797 (2000).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, https://doi.org/10.1093/bioinformatics/btu170 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, https://doi.org/10.1038/nmeth.1923 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297 (2012).
Chen, Y., Lun, A. T. & Smyth, G. K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 5, 1438 (2016).
Spitzer, M., Wildenhain, J., Rappsilber, J. & Tyers, M. BoxPlotR: a web tool for generation of box plots. Nat. Methods 11, 121–122 (2014).
Acknowledgements
Funding was generously provided by The Sea Turtle Conservancy, Florida Sea Turtle Grants Program under project number 17-033R, the Save Our Seas Foundation under project number SOSF 356, the National Save The Sea Turtle Foundation, Inc. under project name Fibropapillomatosis Training and Research Initiative, and a Welsh Government Sêr Cymru II and the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 663830-BU115. This research was also supported by Gumbo Limbo Nature Center, Inc d/b/a Friends of Gumbo Limbo (a 501c3 non-profit organization) through a generous donation through their Graduate Research Grant program and by an Irish Research Council Government of Ireland Postgraduate Scholarship, under project number GOIPG/2020/1056. Warmest thanks to Mark Q. Martindale, Nancy Condron, and the veterinary and rehabilitation staff and volunteers of the Sea Turtle Hospital at Whitney Laboratories. Thanks also are due to David Moraga, Yanping Zhang, and Mei Zhang of UF’s Interdisciplinary Center for Biotechnology Research Core, Whitney Daniel and the staff of the South Carolina Aquarium, Elizabeth Ryan, Brian A. Stacy, and Nicole Stacy for informative discussions, and Florida Fish and Wildlife Conservation Commission’s Meghan Koperski for valuable assistance with permitting.
Author information
Authors and Affiliations
Contributions
D.J.D. designed and supervised the project. J.A.F., D.J.D., K.Y., and L.W. generated the data. K.Y., J.A.F, D.J.D., C.S., P.L. D.R.R., and R.T. performed data and bioinformatics analysis. B.B., D.R.R., C.B.E., and R.T. provided veterinary care, including tumor removal surgeries. D.J.D., J.A.F., K.Y., and L.W. wrote the manuscript. J.W., C.S., and S.C. performed critical reading of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. Additionally, the funding agencies had no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Farrell, J.A., Yetsko, K., Whitmore, L. et al. Environmental DNA monitoring of oncogenic viral shedding and genomic profiling of sea turtle fibropapillomatosis reveals unusual viral dynamics. Commun Biol 4, 565 (2021). https://doi.org/10.1038/s42003-021-02085-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-021-02085-2
This article is cited by
-
Inadvertent human genomic bycatch and intentional capture raise beneficial applications and ethical concerns with environmental DNA
Nature Ecology & Evolution (2023)
-
Preparation of recombinant glycoprotein B (gB) of Chelonid herpesvirus 5 (ChHV5) for antibody production and its application for infection detection in sea turtles
Scientific Reports (2022)
-
Morphologic and physiologic characteristics of green sea turtle (Chelonia mydas) hatchlings in southeastern Florida, USA
Journal of Comparative Physiology B (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.