Abstract
Mitochondrial genes encode key components of the cellular energy machinery, but their genetic analysis is difficult or impossible in most organisms (including plants) because of the lack of viable transformation approaches. We report here a method to block the expression of the mitochondrial nad6 gene encoding a subunit of respiratory complex I in Arabidopsis thaliana, via the modification of the specificity of the RNA-binding protein RNA PROCESSING FACTOR 2 (RPF2). We show that the modified RPF2 binds and specifically induces cleavage of nad6 RNA, almost eliminating expression of the Nad6 protein and consequently complex I accumulation and activity. To our knowledge, this is the first example of a targeted block in expression of a specific mitochondrial transcript by a custom-designed RNA-binding protein. This opens the path to reverse genetics studies on mitochondrial gene functions and leads to potential applications in agriculture.
Similar content being viewed by others
Introduction
Among the few genetic mutations known in plant mitochondria1 are recombinant open reading frames (ORFs) linked to cytoplasmic male sterility (CMS)2,3. CMS is widely used in agriculture to facilitate the production of hybrid lines3. CMS can be suppressed by restorer-of-fertility genes, many of which encode pentatricopeptide repeat (PPR) proteins4,5. PPR proteins are sequence-specific RNA-binding proteins involved in posttranscriptional stages of organelle transcript maturation6. Their modular and predictable interactions with RNA make them amenable to custom modification and design7,8. Angiosperms contain about 500 different PPR separable into two classes (P-class and PLS-class) distinguished by their characteristic patterns of repeated motifs9. Restorer-of-fertility proteins form a clade within the P-class PPR proteins that co-evolves with mitochondrial ORFs causing CMS4. They appear to block expression of specific mitochondrial ORFs by inducing RNA cleavage or degradation, or by preventing translation. The mechanisms by which these processes are achieved are not known in detail5,10. Arabidopsis thaliana has 26 restorer-of-fertility-like (RFL) proteins11, some of which are involved in 5′-end processing of mitochondrial transcripts12,13,14.
The discovery of the PPR code15 describing the correspondence between key residues of PPR motifs and each base of its target RNA8 allows the prediction of a target for a given PPR protein16,17, or inversely the prediction of which PPR protein binds a given target18. It also permits the design of proteins with altered binding capabilities8,15,19,20, suggesting that custom RNA-processing factors with desired targeting specificities could be developed7. In this work, we designed the RFL protein RNA PROCESSING FACTOR 2 (RPF2)13 to bind a new RNA target, located within the coding sequence of nad6, and show that the re-targeting is successful.
Results
Re-design and expression of RPF2
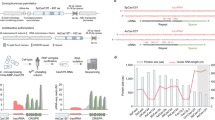
RPF2 (At1g62670) has 16 PPR motifs and 2 known natural targets, located within the 5′-untranslated regions (UTRs) of cox3 and nad913. Using the code defined by Barkan et al.15, RPF2 is predicted to bind to similar sequences 471–455 nt upstream of the cox3 start codon and 301–286 nt upstream of the nad9 start codon (Fig. 1a)21. We searched for other similar sequences in the A. thaliana mitochondrial transcriptome, and found that the sequence at position + 349–365 in the coding sequence of nad6 has only 3 differences to the predicted RPF2-binding site in the cox3 transcript. We modified the coding sequence of RPF2 to incorporate residues that would be predicted to recognize this new target, giving the sequence that we refer to as RPF2-nad6 (Supplementary Fig. 1). RPF2-nad6 was introduced into A. thaliana Col-0 plants via Agrobacterium infection. A similar construct with the native RPF2 cDNA was transformed in parallel as a control. Reverse-transcriptase PCR (RT-PCR) was performed on primary transformants (T1) to ensure that the constructs were integrated in the genome and expressed (Supplementary Fig. 2). The FLAG-tagged protein could not be detected by western blotting in mitochondrial protein extracts, but 12 out of 19 independent transformants carrying the RPF2-nad6 construct displayed slow growth (Fig. 1b), and later in development, dark curled foliage reminiscent of Arabidopsis complex I mutants (Fig. 1c and Supplementary Fig. 3). All control plants (wild type (WT) + native RPF2) were phenotypically indistinguishable from WT (23 independent transformants).
Targeting nad6 with a modified RPF2 protein leads to plants with a slow growth phenotype. a Binding predictions for the RPF2 and RPF2-nad6 proteins on their respective targets (5′-UTRs of cox3 and nad9 for RPF2 and the coding sequence of nad6 for RPF2-nad6). Dark green represents a perfect match, light green a partial match, and magenta a mismatch according to the PPR code15. b, c Phenotypes of plants transformed with native RPF2 or RPF2-nad6 compared with WT. b Four-week-old rosette; c 6-week-old plants grown under 18 h photoperiod
RPF2-nad6 plants have reduced Nad6 and complex I
In order to check whether the phenotypes observed could be linked to an alteration in complex I levels, blue native polyacrylamide gel electrophoresis (BN-PAGE) was performed on isolated leaf mitochondria from T2 plants of the transformants (Fig. 2a). No NADH oxidase activity from complex I was revealed in mitochondria from the slow growing plants bearing the RPF2-nad6 construct. Western blotting of the BN-PAGE gel and probing with an antibody raised against the carbonic anhydrase subunit CA2, one of the first subunits to be incorporated in complex I22, revealed assembled complex I in the WT mitochondria and the RPF2 plants (Fig. 2b) but only a low-molecular-weight subcomplex in RPF2-nad6 mitochondria, suggesting that the membrane arm assembly and subsequent complex assembly were compromised in the latter. This low-molecular-weight sub-complex was not detected in tang2, a PPR mutant altered in the splicing of the nad5 transcript23. These results are consistent with a proposed order of assembly of complex I subunits in which CA2 and Nad2 are involved in the assembly of an initial 200 kDa subcomplex, with Nad6 addition required for the next stage of assembly22,24,25,26. Further analysis of respiratory complex subunits by SDS-PAGE and western blotting (Fig. 2c, Supplementary Fig. 4) showed that Cox2 (complex IV), RISP (complex III), and AtpA (complex V) subunits were unchanged in RPF2-nad6 mitochondria, but Nad9 was decreased and Nad6 was undetectable in mitochondria of RPF2-nad6-transformed plants (Fig. 2c). Very low levels of NDUFS4, a nuclearly encoded subunit of complex I, were detected. Furthermore, alternative oxidase (AOX) was greatly induced in RPF2-nad6 plants (Fig. 2c), as commonly observed for mutants with altered complex I function27,28,29.
RPF2-nad6 plants lack assembled respiratory complex I. a, b Separation of mitochondrial inner membrane protein complexes by blue native PAGE in RPF2-nad6 plants as compared with WT, tang2 (a complex I mutant) and plants transformed with the native RPF2 constructs (RPF2). NADH oxidase activity was revealed in-gel (a), showing the lack of assembled complex I in the RPF2-nad6 plants. A western blotting of a similar gel was probed with an anti-CA2 antibody (b). The black arrow shows a low-molecular-weight assembly intermediate of complex I in the RPF2-nad6 plants. c Western blottings of mitochondrial proteins from RPF2-nad6 plants as compared with WT and control RPF2 plants separated by SDS-PAGE. Uncropped images of the blots are presented in Supplementary Fig. 4
The nad6 transcript is cleaved in RPF2-nad6 plants
A quantitative analysis of the expression levels of mitochondrial transcripts in the modified RPF2 plants as compared with WT was obtained by RNA sequencing (RNA-seq). This analysis revealed that levels of the nad6 transcript were reduced about fourfold in RPF2-nad6 (Fig. 3a). An explanation for this reduction was sought through northern blotting with nad6-specific probes. Leaf and flower RNAs were probed with biotinylated oligonucleotide probes targeted to the vicinity of the predicted RPF2-nad6 binding site (Fig. 3b). Probe 364 AS (covering the predicted binding site) hybridized to a single ~780 nt transcript in WT samples corresponding to the expected size of the mature nad6 transcript (179 nt 5′-UTR followed by 601 nt of coding sequence—the nad6 mRNA in A. thaliana has no stop codon or 3′-UTR30). In the RPF2-nad6 plants, this hybridization signal was much weaker and accompanied by a more intense signal migrating at ~500–600 nt, suggesting that the nad6 transcript is cleaved in the RPF2-nad6 plants. With the 556 AS probe (hybridizing 3′ of the predicted binding site), a second potential cleavage product was detected, migrating at ~200–250 nt.
The nad6 transcript is cleaved in the plants transformed with the RPF2-nad6 constructs. a The plot shows the relative expression of all mitochondrial transcripts in RPF2-nad6 plants compared with WT as log2 ratios of read counts. Each point represents one of four biological replicates. The reduced expression of nad6 in RPF2-nad6 plants is significant (FDR-corrected p-value 6.3 × 10−78; Wald’s test using DESeq242). b Northern blottings of leaf and flower RNAs from two independent RPF2-nad6 transformants and WT plants (top panels). The probe 364 AS covers the predicted RPF2-nad6-binding site and 556 AS hybridizes 3′ of the binding site. The lower panels show the gels stained with ethidium bromide before transfer to control for loading differences. The sizes on the left indicate estimated sizes of the hybridizing RNAs, sizes on the right are markers
To map the exact site of the apparent cleavage, we performed 5′-rapid amplification of cDNA end (RACE) PCR and circular RT-PCR (cRT-PCR) experiments. The sizes of the 5′-RACE and cRT-PCR products were determined by migration on a 3% low melting agarose gel and sequencing of cloned products (Supplementary Fig. 5). Multiple transcript ends could be mapped within a region encompassing 55 nucleotides beginning at or near the end of the predicted RPF2-nad6-binding site (Fig. 4). The cRT-PCR revealed that the cleaved RNA is lacking a segment of ~33 nt. This could result either from a double endonucleolytic cleavage or a single cleavage followed by exonucleolytic trimming (Supplementary Fig. 6). We found no evidence for the additional product expected from a double cleavage using RACE with primers targeted within this segment, or by sequencing small (–17-50 nt) RNAs prepared from WT and RPF2-nad6 plants (Supplementary Fig. 7). The 5′–3′ exonuclease activity is unknown from plant mitochondria31; thus, we suggest that the initial cleavage is 55 nt 3′ of the RFP-nad6-binding site, followed by 3′–5′ exonucleolytic degradation, which is inhibited from extending further by the bound PPR protein. This scenario is supported by the observation of un-templated A addition at the 3′-end of the 5′-cleavage product (Supplementary Fig. 5), known to induce 3′–5′ exonuclease activity by polynucleotide phosphorylase32.
Recapitulative map of nad6 transcript cleavage in RPF2-nad6 plants using the 5′-RACE and cRT-PCR results. The coordinates of this region on the Col-0 mitochondrial genome are 165,200 to 165,300. The predicted RPF2-nad6 binding (165,221–165,236) and cleavage (165,269) sites are highlighted. The black and yellow triangles indicate the 5′-ends of the cleaved products as determined by cRT-PCR and 5′-RACE PCR, respectively. White triangles show the 3′-ends of the cleaved products as determined by cRT-PCR. The figures near the triangles indicate the numbers of clones obtained
Off-target cleavage of rps3-rpl16 transcripts
The cleavage of nad6 transcripts in close proximity to the predicted RPF2-nad6 binding site and the mapping of an RNA end at this site strongly suggests that RPF2-nad6 does indeed recognize the target sequence it was designed to bind. Although the RNA-seq data (Fig. 3a) did not provide obvious evidence of loss of any other transcripts besides nad6, this does not rule out more subtle off-target effects. We searched for other potential targets in the mitochondrial transcriptome with the pattern ‘NAAAURCGACCUNUCY’ (based on the nad6 target site, but allowing redundancy where the natural targets of RPF2 suggest base-specificity is less than perfect). A single sequence matches exactly (the nad6 site), but a sequence in the rps3 coding sequence contains only one minor (C–U) mismatch to this pattern, and as distinction between C and U appears to be the most difficult for PPR motifs19, we investigated potential cleavages in the vicinity of this site (Supplementary Fig. 8). We discovered that the rps3-rpl16 co-transcript is cleaved ~20 nt 3′ of the predicted binding site, leading to accumulation of the 3′ end of the transcript but loss of the 5′-part; expression of Rps3 and Rpl16 is presumably unaffected because the full-length transcript containing both overlapping reading frames is present at WT levels in the RPF2-nad6 plants. Thus, as seen for natural RPF2, RPF2-nad6 can tolerate mismatches to its predicted preferred target sequence, leading to ‘off-target’ RNA cleavage.
The endonuclease that cleaves remains unidentified
An interesting feature of the cleavages induced by RPF2-nad6 is their distance from the predicted binding sites. This “cleavage at a distance” is also inferred to occur with the RNAs that are natural targets of unmodified RPF2. The predicted binding sites on nad9 and cox3 are, respectively, 83 nt and 77 nt 5′ of the final mRNA termini in Col-0 (Supplementary Fig. 9). Thus, identical (or near-identical) proteins apparently induce cleavages at different distances on different RNAs. What might be the mechanism? The relatively long distances to the proposed cleavage sites suggest the involvement of another factor, and/or RNA secondary structure. The tRNA processing enzyme proteinaceous RNAse P (PRORP)33 was implicated in the RFL2-promoted cleavage of orf29110 and is proposed to cleave nad6 mRNA 17 nt upstream of its stop codon34. In both cases, tRNA-like secondary structures can form that are a substrate for PRORP35. We could find no such structure in the vicinity of the RPF2-nad6 cleavage sites or near the RPF2 cleavage sites in nad9 and cox3 (Supplementary Fig. 10), making it unlikely that PRORP is the endonuclease in these cases. We also conclude from these folding simulations that binding of RPF2 or RPF2-nad6 does not obviously alter the structure of the RNA around the cleavage sites. The endonucleases MNU1 and MNU2 have been proposed to act in concert with RPF2 to cleave nad9 and cox336, but we still observed RPF2-nad6-induced cleavage of nad6 in a double mnu1 mnu2 mutant background (Supplementary Fig. 11, 12). The identity of the endonuclease presumed to be working with RFP2-nad6 is thus still obscure.
Discussion
We have shown that it is possible to redirect an RFL-type PPR protein to a new target within mitochondria in vivo and effectively block expression of the target transcript via induced cleavage of the mRNA and a resulting loss of the translation product. To our knowledge, it is the first time a modified PPR protein has been used to block the expression of an organellar transcript in this way. A previous attempt to achieve a similar result, the directed knockdown of matR, the mitochondrial maturase whose gene lies within nad1 intron 4, used synthetic ribozymes37. The knockdown effect yielded by this technique was at best a 50% reduction in the transcript and protein, much less than the fourfold decrease in transcript levels and almost total lack of Nad6 protein achieved in this work. We believe that the approach we demonstrate here should provide a general means to implement reverse genetics approaches in plant mitochondria given the ease with which PPR proteins can be designed to recognize different target sequences7,20. Extension of this approach to plastids is possible in principle, but will probably require the addition of an endonuclease domain to the PPR; there is no evidence so far that RFL proteins can induce cleavage of plastid RNAs. Lack of specificity (particularly distinction between C and U) remains an issue to be solved. One possible approach may be to reduce the length of the PPR tract; experiments with synthetic PPR proteins suggest that 10 PPR motifs are sufficient for optimal binding, whereas 14 motifs allow the protein-RNA complex to tolerate more mismatches19. Natural PPR proteins are often longer than this and can bind a range of similar sequences, as demonstrated for the maize plastid protein PPR1038. Natural RFL proteins contain up to 15–20 PPR motifs4,11, implying a similar ability to bind a range of related targets, in accordance with the observed ability of RPF2 and RPF2-nad6 (16 motifs) to bind two closely related sequences each.
Methods
Cloning
The synthetic RPF2-nad6 gene as well as a fragment containing the NOS promoter, the 25 amino acid Solanum tuberosum formate dehydrogenase (FDH) targeting peptide and a FLAG tag were commercially synthesized, digested with BspHI and BamHI, and EcoRI and BspHI, respectively, and cloned together into the pCAMBIA1380 binary vector39. The native RPF2 gene was amplified from Col-0 genomic DNA with the Takara PrimeSTAR HS polymerase using the primers RPF2pETRcaF and RPF2pETBamH1R (Supplementary Table 1) and cloned with the same promoter-targeting sequence and FLAG cassette in pCAMBIA1380 (Supplementary Fig. 1a). The synthetic genes were transferred to Agrobacterium tumefaciens and introduced into A. thaliana plants by floral dip40. In short, floral stems carrying unpollinated buds were dipped for 20–30 s into a fresh culture of A. tumefaciens resuspended in a 10% sucrose solution containing 0.5 μL mL−1 Silwett. The plants were covered and kept under low light for 24 h, staked, and grown until the siliques were ready to harvest. The transformants were selected on 25 μg mL−1 hygromycin.
RNA sequencing
Total RNA was isolated from 6-week-old rosette leaves and bolting flower buds of Col-0 and one of the RPF2-nad6 transformed lines (T2) with an RNeasy Qiagen kit. Four independent libraries for each genotype were made from 500 ng of Turbo DNase (Ambion) treated total RNA using an Illumina TruSeq Stranded kit. The sequencing run (MiSeq Reagent Kit v3, 150 cycles) was performed on an Illumina MiSeq sequencer. Reads were mapped to the Col-0 mitochondrial transcriptome (derived from accession JF729201) with Salmon41 v0.11.2 and the count data analyzed with DESeq242 v1.20.0.
Northern blotting
RNA from rosette leaves and flowers was extracted using PureZol (Bio-Rad). Eight micrograms of total RNA were run on a 1.2 % denaturing agarose gel and transferred onto Hybond N + membrane (Amersham). Northern blotting was performed as described previously43 using oligonucleotide probes labeled in 5′ with biotin (Supplementary Table 1). The membranes were pre-hybridized for 1–2 h at 50 °C in 5 × SSC, 7% SDS, 100 μg ml−1 heparin, 20 mM Na2HPO4 pH 7.5 and hybridized overnight in the same buffer containing 1 nM biotinylated probe. Three short washes in 3 × SSC, 5% SDS, 25 mM Na2HPO4 pH 7.5 were performed at room temperature. The northern blottings were developed with the Pierce Chemiluminescent Nucleic Acid Detection Module Kit.
5′-RACE PCR
5′-RACE was performed using the SMARTer RACE cDNA Amplification Kit (Clontech) according to the manufacturer’s instructions. Random hexamers were used to prime reverse transcription. The PCR amplification was done using a specific primer in the nad6 coding sequence (nad6-554Rv, Supplementary Table 1). The RACE PCR products obtained for the Col-0 and RPF2-nad6 plants were run on a 3% low melting agarose gel (Supplementary Fig. 5), cloned into pGEM-T Easy (Promega) and sequenced.
Circular RT-PCR
Crude preparations of mitochondria were isolated from 4-week-old Col-0 and RPF2-nad6 seedlings grown on half-strength Murashige and Skoog medium. Mitochondrial RNA was purified from mitochondrial pellets using PureZol (Bio-Rad) and for each reaction, 2–3 μg were treated with Turbo DNase (Ambion). RNA was circularized using the T4 RNA ligase and reverse transcription was performed with the Superscript III (Invitrogen) using specific primers (nad6 cRT 500R for the fragment downstream of the cleavage site and nad6 202R for the fragment upstream of it). PCR was performed with the same reverse primer (or a nested primer, nad6 149R) and a specific forward primer (nad6 517F and nad6 500R or nad6 244F and nad6 202R or nad6 149R). PCR products were cloned into pGEM-T Easy and sequenced. A similar reverse transcription was performed to check the ends of the rps3-rpl16 transcript using the primers rpl16-cRT-M4 and rps3-cRT-R2. Primer sequences are presented in Supplementary Table 1.
Protein electrophoresis and western blotting
BN-PAGE, NADH oxidase activity staining and western blotting were performed as previously described16. The blots were probed using antibodies against Nad944 (1:50,000 dilution), Nad645 (AS15 2926 from Agrisera at 1:1000 dilution), NDUFS446 (1:5000 dilution), and CA247 (1:2500 dilution) from complex I, the Rieske iron–sulfur protein of complex III48 (RISP, 1:5000 dilution), Cox2 of complex IV (AS04 053A from Agrisera at 1:5000 dilution), and NDB2 of the external NADH dehydrogenase49 (1:5000 dilution). The antibodies against AtpA of the ATP synthase (1:1000 dilution), the AOX50 (1:1000 dilution), and the mitochondrial membrane Porin (1:5000 dilution) were provided by Dr T. Elthon (School of Biological Sciences, University of Nebraska).
Data availability
The datasets generated in this study have been deposited in the NCBI SRA database as accession SRP158968 (accession numbers SRR7760264–SRR7760269 for the sRNA-seq data and SRR7760270–SRR7760277 for the RNA-seq data).
References
Kubo, T. & Newton, K. J. Angiosperm mitochondrial genomes and mutations. Mitochondrion 8, 5–14 (2008).
Schnable, P. S. & Wise, R. P. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180 (1998).
Chase, C. D. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23, 81–90 (2007).
Fujii, S., Bond, C. S. & Small, I. D. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl Acad. Sci. USA 108, 1723–1728 (2011).
Gaborieau, L., Brown, G. G. & Mireau, H. The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility. Front. Plant Sci. 7, 1816 (2016).
Barkan, A. & Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442 (2014).
Yagi, Y., Nakamura, T. & Small, I. The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 78, 772–782 (2014).
Shen, C. et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 7, 11285 (2016).
Cheng, S. et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547 (2016).
Fujii, S. et al. The Restorer-of-fertility-like 2 pentatricopeptide repeat protein and RNase P are required for the processing of mitochondrial orf291 RNA in Arabidopsis. Plant J. 86, 504–513 (2016).
Dahan, J. & Mireau, H. The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol. 10, 1469–1476 (2013).
Jonietz, C., Forner, J., Hildebrandt, T. & Binder, S. RNA PROCESSING FACTOR3 is crucial for the accumulation of mature ccmC transcripts in mitochondria of Arabidopsis accession Columbia. Plant Physiol. 157, 1430–1439 (2011).
Jonietz, C., Forner, J., Hölzle, A., Thuss, S. & Binder, S. RNA PROCESSING FACTOR2 is required for 5′ end processing of nad9 and cox3 mRNAs in mitochondria of Arabidopsis thaliana. Plant Cell 22, 443–453 (2010).
Hölzle, A. et al. A restorer of fertility-like PPR gene is required for 5′-end processing of the nad4 mRNA in mitochondria of Arabidopsis thaliana. Plant J. 65, 737–744 (2011).
Barkan, A. et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8, e1002910 (2012). https://doi.org/10.1371/journal.pgen.1002910.
Lee, K. et al. The mitochondrial pentatricopeptide repeat protein PPR19 is involved in the stabilization of NADH dehydrogenase 1 transcripts and is crucial for mitochondrial function and Arabidopsis thaliana development. New Phytol. 215, 202–216 (2017).
Wang, C. et al. The pentatricopeptide repeat protein MTSF2 stabilizes anad1 precursor transcript and defines the 3′ end of its 5′-half intron. Nucleic Acids Res. 45, 6119–6134 (2017).
Yap, A. et al. AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J. 81, 661–669 (2015).
Miranda, R. G., McDermott, J. J. & Barkan, A. RNA-binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR–RNA interactions. Nucleic Acids Res. 46, 2613–2623 (2018).
Coquille, S. et al. An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 5, 5729 (2014).
Binder, S., Stoll, K. & Stoll, B. P-class pentatricopeptide repeat proteins are required for efficient 5′ end formation of plant mitochondrial transcripts. RNA Biol. 10, 1511–1519 (2013).
Meyer, E. H., Solheim, C., Tanz, S. K., Bonnard, G. & Millar, A. H. Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. J. Biol. Chem. 286, 26081–26092 (2011).
Colas des Francs-Small, C. et al. The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiol. 165, 1409–1416 (2014).
Braun, H.-P. et al. The life of plant mitochondrial complex I. Mitochondrion 19, 295–313 (2014).
Schimmeyer, J., Bock, R. & Meyer, E. H. L-Galactono-1,4-lactone dehydrogenase is an assembly factor of the membrane arm of mitochondrial complex I in Arabidopsis. Plant Mol. Biol. 90, 117–126 (2016).
Meyer, E. H. Proteomic investigations of complex I composition: how to define a subunit? Front. Plant Sci. 3, 106 (2012).
Haïli, N. et al. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 41, 6650–6663 (2013).
Zmudjak, M. et al. mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. New Phytol. 199, 379–394 (2013).
Cohen, S. et al. nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 78, 253–268 (2014).
Raczynska, K. D. et al. Plant mitochondrial genes can be expressed from mRNAs lacking stop codons. FEBS Lett. 580, 5641–5646 (2006).
Ruwe, H., Wang, G., Gusewski, S. & Schmitz-Linneweber, C. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 44, 7406–7417 (2016).
Holec, S. et al. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell. Biol. 26, 2869–2876 (2006).
Gobert, A. et al. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 17, 740 (2010).
Forner, J., Weber, B., Thuss, S., Wildum, S. & Binder, S. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 35, 3676–3692 (2007).
Gobert, A. et al. Structural insights into protein-only RNase P complexed with tRNA. Nat. Commun. 4, 1353 (2013).
Stoll, B. & Binder, S. Two NYN domain containing putative nucleases are involved in transcript maturation in Arabidopsis mitochondria. Plant J. 85, 278–288 (2016).
Sultan, L. D. et al. The reverse transcriptase/RNA maturase protein MatR is required for the splicing of various group II Introns in Brassicaceae mitochondria. Plant Cell 28, 2805–2829 (2016).
Miranda, R. G., Rojas, M., Montgomery, M. P. & Gribbin, K. P. RNA binding specificity landscape of the pentatricopeptide repeat protein PPR10. RNA 23, 586–599 (2017).
Bevan, M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721 (1984).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Falcon de Longevialle, A. et al. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 56, 157–168 (2008).
Lamattina, L., Gonzalez, D., Gualberto, J. & Grienenberger, J. M. Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur. J. Biochem. 217, 831–838 (1993).
Koprivova, A. et al. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. J. Biol. Chem. 285, 32192–32199 (2010).
Meyer, E. H. et al. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 151, 603–619 (2009).
Perales, M. et al. Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I + III2 levels and alters mitochondrial physiology in Arabidopsis. J. Mol. Biol. 350, 263–277 (2005).
Carrie, C. et al. Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J. Biol. Chem. 285, 36138–36148 (2010).
Carrie, C. et al. Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 582, 3073–3079 (2008).
Elthon, T. E., Nickels, R. L. & McIntosh, L. Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 89, 1311–1317 (1989).
Acknowledgements
This work was funded by the Australian Research Council (CE140100008, FL140100179).
Author information
Authors and Affiliations
Contributions
C.C.d.F.-S. and I.S. conceived and designed the experiments. C.C.d.F.-S. and L.V.P.S carried out the experiments. C.C.d.F.-S., L.V.P.S., and I.S. analyzed the data, prepared the figures, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Colas des Francs-Small, C., Vincis Pereira Sanglard, L. & Small, I. Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun Biol 1, 166 (2018). https://doi.org/10.1038/s42003-018-0166-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-018-0166-8
This article is cited by
-
The genetic basis of cytoplasmic male sterility and fertility restoration in wheat
Nature Communications (2021)
-
Expanding the binding specificity for RNA recognition by a PUF domain
Nature Communications (2021)
-
Engineered PPR proteins as inducible switches to activate the expression of chloroplast transgenes
Nature Plants (2019)
-
Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing
Nature Plants (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.