Abstract
Mycobacterium saskatchewanense is a species of pigmented slow-growing Non-Tuberculous Mycobacteria (NTM), positive for Mycobacterium avium complex (MAC) by AccuProbe system. MAC organisms have frequently been isolated from different medical devices. This is the first study reporting isolation of M. saskatchewanense from medical devices and highlights the importance of correctly identifying the NTMs that often colonize sanitary water. GenoType Mycobacterium CM CE-IVD kit (CM) was used as the first step of NTM strain identification, and all positive cultures were found to be components of MAC. Then, GenoType NTM-DR CE-IVD kit (NTM-DR) was used to differentiate the different species. Sub-culture on solid media were used for: (i) phenotypical confirmation by colony morphology and Matrix-Assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF) mass spectrometry; (ii) molecular confirmation by Next Generation Sequencing. All positive cultures were identified as M. intracellulare by CM and NTM-DR assays, whereas colony morphology showed bright yellow scotochromogenic growth. MALDI-TOF analyses identified the strains as M. saskatchewanense with a high score, and identification was confirmed by NGS analysis based on the hsp-65 region. This paper suggests that it is important to actively monitor NTM contamination in medical devices that use sanitary water, to prevent the possibility of patients becoming infected.
Similar content being viewed by others
Introduction
Non-Tuberculous Mycobacteria (NTM) are a large and heterogeneous group of acid-fast bacilli that are widespread in the environment. Their cell walls contain a high proportion of fatty acids, making them highly hydrophobic. This enables mycobacteria to form biofilm and survive chemical treatments used to disinfect water, such as chlorine. For these reasons, NTM can cause water-borne infections, and healthcare facility water systems can be a reservoir for these microorganisms1,2,3,4.
Mycobacterium Avium Complex (MAC) organisms, including M. avium, M. intracellulare and M. chimaera, are the most frequent NTM causing chronic and disseminated disease in immune-compromised patients. MAC organisms have frequently been isolated from water samples, collected from different medical devices. In particular, in 2013 M. chimaera was identified as the causative agent of invasive infections in cardiothoracic surgery patients with extracorporeal circulation. In 2015 it emerged that the heater-cooler units (HCU) used during open-chest heart surgery were contaminated with M. chimaera causing infection by airborne transmission through ventilation fans5,6.
M. saskatchewanense has been identified as a new species of pigmented and slow-growing NTM, positive for Mycobacterium avium complex (MAC) by AccuProbe system7. Molecular assays based on DNA STRIP technology are the most commonly used tests to identify clinically relevant mycobacteria (approximately 30 species) from cultured material. However, these rapid assays may mis-identify NTM species. Currently, microbiology laboratories can identify most NTM species (approximately 180 species) either through mass spectrometry, which often requires a subculture on solid media in order to obtain a high level of confidence, or through NGS analysis requiring a specialized technician and expensive equipment. This study describes the first isolation of M. saskatchewanense from medical devices and highlights the importance of correctly identifying NTMs that often colonize sanitary water.
Results
Sixteen (13%) out of 112 dialysis fluid MGIT cultures were positive for NTM, verified by acid-fast bacilli staining. All positive cultures were identified as M. intracellulare by Genotype CM and NTM-DR molecular assays. However, sub-cultures on Middlebrook 7H11 agar did not produce the colony morphology expected for M. intracellulare, as bright yellow scotochromogenic growth was observed (Fig. 1). Further analyses performed by Matrix-Assisted Laser Desorption/Ionization-Time of Flight” (MALDI-TOF) mass spectrometry identified all the strains as M. saskatchewanense with a high score (> 1.8).
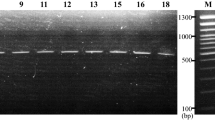
The result obtained by hsp65-based sequencing with Deeplex Myc-TB test, confirmed the identity of the bacterial isolates as M. saskatchewanense (Fig. 2).
Discussion
Nontuberculous mycobacteria (NTM) are frequently the cause of opportunistic infection in immunocompromised hosts. Their ability to form biofilm and withstand chemical treatments has caused them to emerge as opportunistic pathogens within healthcare facilities1.
Numerous studies have reported elevated concentrations of mycobacteria in water systems within healthcare facilities8,9,10. Plumbing systems of large structures often have dead legs and dead ends, which may harbour biofilms, potentially including NTM. Furthermore, there is substantial evidence that the zinc in galvanized plumbing may contribute to the persistence of MAC organisms in these distribution systems11.
Waterborne infections from nontuberculous mycobacteria in healthcare facility water systems have been systematically reviewed. The most commonly affected patient populations were immunocompromised, post-surgical, and haemodialysis. The main routes of exposure included central venous catheters (CVCs), wound exposure, and contamination during surgery1,4.
Contamination of medical devices or aqueous solutions is often implicated in the transmission of infection. Recently, the slow grower M. chimaera has been reported in the setting of contaminated heater-cooler devices used in open-heart surgery and extracorporeal membrane oxygenation procedures5,12.
Our previous research showed that M. chimaera subtypes circulating in hospital plumbing systems passed through absolute filters, highlighting the possibility that other medical equipment using sanitized water, such as endoscope reprocessing devices or haemodialysis systems, could also become contaminated by NTM13.
So far in haemodialysis patients a few outbreaks have been reported, due to contamination by NTM stains belonging to M. chelonae and to M. abscessus complex and inadequate disinfection of disposable high-flux hollow-firer dialyzers14. In contrast, peritonitis caused by NTM is an important complication in peritoneal dialysis patients. According to the review by Song et al.15, over half of the peritonitis cases could be attributed to rapidly growing Mycobacterium such as M. fortuitum (38.6%) and M. chelonae (14.0%). The prevalence, risk factors, and mortality of NTM infections in patients with end-stage renal disease have recently been examined, revealing an increased risk of mortality in cases involving NTM diagnosis16.
M. saskatchewanense was first identified in 2004, as a new species of pigmented slow-growing NTM, positive for Mycobacterium avium complex (MAC) by AccuProbe system from sputum and pleural fluid of a patient with bronchiectasis7. A further report associated M. saskatchewanense with a chronic kidney disease patient, who required dialysis for 2 years prior to evaluation for renal transplant17.
This is the first paper where M. saskatchewanense has been isolated from a large number of ultrapure dialysis fluid water samples.
Differentiation of mycobacteria to the species level by genotypic and phenotypic assay sometimes leads to inaccurate results. Here we compared the identification methods commonly used in mycobacteriology laboratories: DNA STRIP technology, MALDI-TOF mass spectrometry, and hsp65-based sequencing.
The correct identification of M. saskatchewanense was obtained by MALDI-TOF mass spectrometry with a high score, while DNA STRIP technology, the most commonly used method for NTM identification, misidentified it as M. intracellulare. Culture remains essential for NTM isolation and colony morphology can support identification. Therefore, laboratories that use DNA STRIP technology for NTM identification must use a confirmatory method to achieve correct classification.
This paper suggests that it is important to actively monitor NTM contamination in medical devices that use sanitary water, to prevent the possibility of patients becoming infected.
Methods
From August to November 2022, 112 ultrapure dialysis fluid samples from dialysis machines were processed by the referral centre for the detection of mycobacteria from environmental specimens in the Emilia-Romagna Region, at the Microbiology Unit, IRCCS University Hospital of Bologna, Italy.
Each water sample (1 L) was filtered with cellulose nitrate membranes (0.45 µm), using Microsart filtration system (Sartorius, Germany) according to ECDC guidelines and the residue resuspended in 10 mL of 0.9% saline solution12. Concentrated samples were treated as previously described13. Samples were decontaminated using BBL MycoPrep solution (Becton Dickinson, USA), resuspended in 2 mL of phosphate buffered solution, inoculated onto solid medium (Lowenstein Jensen, Heipha Diagnostics, Germany) and into Middlebrook 7H9 Broth (MGIT, Becton Dickinson, USA) and cultured for 42 days. Ziehl–Neelsen staining was used to confirm mycobacterial presence in positive cultures.
Mycobacteria from positive MGIT cultures were identified as NTM by DNA STRIP technology using first Genotype CM CE-IVD kit (CM, Bruker, Germany) and then, if belonging to Mycobacterium avium complex, analysed by Genotype NTM-DR CE-IVD kit (NTM-DR, Bruker, Germany). Positive MGIT were sub-cultured in Middlebrook 7H11 Agar (7H11 plate, Becton Dickinson, USA) for approximately 2 weeks to obtain microbial growth for phenotypic confirmation by morphological features and MALDI-TOF mass spectrometry (Bruker, Germany). A small amount of bacterial biomass, picked from 7H11 plates, was extracted by adding 50 µL of pure acetonitrile, 50 µL of 70% formic acid and vortexing. Then, the solution was centrifuged and 1 μl of supernatant was spotted on a MALDI target, and allowed to air dry before MALDI-TOF analysis.
Bacterial strains were subsequently sequenced with Deeplex Myc-TB CE-IVD kit (GenoScreen, France) according to the manifacturer’s instructions, to confirm identification.
Data availability
The dataset used during the current study is available from the corresponding author on reasonable request.
References
Li, T., Abebe, L. S., Cronk, R. & Bartram, J. A systematic review of waterborne infections from nontuberculous mycobacteria in health care facility water systems. Int. J. Hyg. Environ. Health. 220(3), 611–620. https://doi.org/10.1016/j.ijheh.2016.12.002 (2017).
Cervia, J. S., Ortolano, G. A. & Canonica, F. P. Hospital tap water: A reservoir of risk for health care-associated infection. Infect. Dis. Clin. Pract. 16(6), 349–353. https://doi.org/10.1097/IPC.0b013e318181fa5e (2008).
Donohue, M. J. et al. Increased frequency of nontuberculous mycobacteria detection at potable water taps within the United States. Environ. Sci. Technol. 49(10), 6127–6133. https://doi.org/10.1021/acs.est.5b00496 (2015).
Desai, A. N. & Hurtado, R. M. Infections and outbreaks of nontuberculous mycobacteria in hospital settings. Curr. Treat. Options Infect. Dis. 10(2), 169–181. https://doi.org/10.1007/s40506-018-0165-9 (2018).
European Centre for Disease Prevention and Control. Invasive cardiovascular infection by Mycobacterium chimaera associated with 3T heater-cooler system used during open-heart surgery – 18 November 2016. Stockholm: ECDC; 2016 Available from: https://www.ecdc.europa.eu/sites/default/files/media/en/publications/Publications/RRA-mycobacterium-chimaera-November-2016.pdf
Ghodousi, A. et al. Genomic analysis of cardiac surgery-associated Mycobacterium chimaera infections in Italy. PLoS One 15(9), e0239273. https://doi.org/10.1371/journal.pone.0239273 (2020).
Turenne, C. Y. et al. Mycobacterium saskatchewanense sp. Nov., a novel slowly growing scotochromogenic species from human clinical isolates related to Mycobacterium interjectum and Accuprobe-positive for Mycobacterium avium complex. Int. J. Syst. Evol. Microbiol. 54(3), 659–667. https://doi.org/10.1099/ijs.0.02739-0 (2004).
Covert, T. C., Rodgers, M. R., Reyes, A. L. & Stelma, G. N. Jr. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65(6), 2492–2496. https://doi.org/10.1128/AEM.65.6.2492-2496.1999 (1999).
du Moulin, G. C., Stottmeier, K. D., Pelletier, P. A., Tsang, A. Y. & Hedley-Whyte, J. Concentration of Mycobacterium avium by hospital hot water systems. JAMA 260, 1599–1601. https://doi.org/10.1001/jama.260.11.1599 (1988).
Peters, M. et al. Isolation of atypical mycobacteria from tap water in hospitals and homes: Is this a possible source of disseminated MAC infection in AIDS patients?. J. Infect. 31(1), 39–44. https://doi.org/10.1016/S0163-4453(95)91333-5 (1995).
Falkinham, J. O. 3rd. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9(2), 177–215. https://doi.org/10.1128/CMR.9.2.177 (1996).
Eu protocol for M. chimaera, Technical document; Stockholm: ECDC; 2015 available at: https://www.ecdc.europa.eu/en/publications-data/eu-protocol-case-detection-laboratory-diagnosis-and-environmental-testing
Bisognin, F. et al. Investigating the origin of Mycobacterium chimaera contamination in heater-cooler units: Integrated analysis with Fourier transform infrared spectroscopy and whole-genome sequencing. Microbiol. Spectr. 10(6), e0289322. https://doi.org/10.1128/spectrum.02893-22 (2022).
Lowry, P.W. et al. Mycobacterium chelonae infection among patients receiving high-flux dialysis in a hemodialysis clinic in California. J. Infect. Dis. 161 (1), 85–90 (1990). Published by: Oxford University Press Stable URL: https://www.jstor.org/stable/30119628
Song, Y. et al. Peritoneal dialysis-associated nontuberculous mycobacterium peritonitis: A systematic review of reported cases. Nephrol. Dial. Transplant. 27(4), 1639–1644. https://doi.org/10.1093/ndt/gfr504 (2012).
Toth, E. et al. Non-tuberculous mycobacterial infections in patients with end-stage renal disease: Prevalence, risk factors, and mortality. J. Investig. Med. Online ahead of print. Oct 6:jim-2022–002462. (2022). https://doi.org/10.1136/jim-2022-002462
Di Mento, G. et al. Mycobacterium saskatchewanense strain associated with a chronic kidney disease patient in an Italian transplantation hospital and almost misdiagnosed as Mycobacterium tuberculosis. Infect. Control Hosp. Epidemiol. 40(4), 496–497. https://doi.org/10.1017/ice.2019.6 (2019).
Acknowledgements
We acknowledge the excellent technical assistance provided by Eleonora Gatti. We thank Jackie Leeder, BSc, for English language editing.
Author information
Authors and Affiliations
Contributions
F.B. and P.D.M. conceived and designed the experiments; V.F. and F.S. performed the experiments and curated the data; F.B., V.F., G.L. and P.D.M. analyzed the data; F.B., G.L. and P.D.M. wrote and revised the paper; T.L. and P.D.M. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bisognin, F., Ferraro, V., Sorella, F. et al. First isolation of Mycobacterium saskatchewanense from medical devices. Sci Rep 13, 21628 (2023). https://doi.org/10.1038/s41598-023-48974-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-48974-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.