Abstract
Disruption of intestinal epithelial functions is linked to Crohn disease (CD) pathogenesis. We identified a widespread reduction in the expression of long non-coding RNAs (lncRNAs) including LHFPL3-AS2 in the treatment-naïve CD ileum of the RISK pediatric cohort. We validated the reduction of LHFPL3-AS2 in adult CD and noted a further reduction in patients with more severe CD from the RISK cohort. LHFPL3-AS2 knockdown in Caco-2 cells robustly affected epithelial monolayer morphogenesis with markedly reduced confluency and spreading, showing atypical rounding, and clumping. mRNA-seq analysis of LHFPL3-AS2 knockdown cells highlighted the reduction of genes and pathways linked with apical polarity, actin bundles, morphogenesis, and the b-catenin-TCF4 complex. LHFPL3-AS2 knockdown significantly reduced the ability of cells to form an internal lumen within the 3-dimensional (3D) cyst model, with mislocalization of actin and adherent and tight junction proteins, affecting epithelial polarity. LHFPL3-AS2 knockdown also resulted in defective mitotic spindle formation and consequent reduction in epithelial proliferation. Altogether, we show that LHFPL3-AS2 reduction affects epithelial morphogenesis, polarity, mitotic spindle formation, and proliferation, which are key processes in maintaining epithelial homeostasis in CD. Reduced expression of LHFPL3-AS2 in CD patients and its further reduction with ileal ulceration outcome, emphasizes its significance in this context.
Similar content being viewed by others
Introduction
The intestinal epithelium serves as the first line of defense between the luminal microbiota and the host immune cells. The establishment of the apicobasal epithelial barrier polarity is dependent upon fine regulation of the cytoskeletal network, the secretory organelles (ER, Golgi)1, 2, and the intracellular trafficking machinery3. The three types of cell polarity include the arrangement of molecules through the XY axis, the distribution of molecules around the cell (radial polarity), and the apicobasal polarity on the Z axis4. Intracellular trafficking and sorting support the distribution of the polarity domains5, 6. Adhesion molecules (cadherins) and tight junction (TJ) proteins establish cellular connections and restrict the diffusion of protein between the apical and basal cell membrane domains7. Polarized actin and microtubule cytoskeleton serve as rails for intracellular transport8,9,10 and for orienting organelle positioning, a process that is also required during mitotic spindle formation and cell division2. Highly conserved polarity complexes were described; the Crumbs/Pals1/PatJ which mostly localized apically, the Par3/Par6/aPKC/CDC42 that localized at the TJs and is linked with the bipolar attachment of spindle microtubules to kinetochores in metaphase11 and is involved in actin cytoskeleton re-organization during cell shape changes12, and Scribble (SCRIB))/Dlg/Lgl, which localized at the basolateral surface4. Additionally, RAB17 was shown to regulate intracellular membrane trafficking in polarized epithelial cells by recycling endosomes13. Failure in sorting and trafficking leads to microvillus inclusion disease, which is linked with severe diarrhea and the inability to absorb nutrients14. The disruption of epithelial barrier functions and polarity contributes to abnormal epithelial functions also related to Crohn disease (CD) pathogenesis3,15.
In our previous publication, we identified a widespread reduction in the expression of epithelial long non-coding RNAs (lncRNAs) in treatment naïve CD ileum, including a significant reduction in LHFPL3-AS216, a novel relatively uncharacterized lncRNA. LncRNAs are known to regulate fundamental cellular processes, including gene expression, regeneration, and proliferation. This analysis used mRNAseq selection and was limited in its ability to capture non-poly adenylated RNAs17. Several lncRNAs are also suggested to regulate epithelial cytoskeleton function18, 19 and to modulate critical cell cycle regulators including cyclins, cyclin-dependent kinases (CDKs), and CDK inhibitors20,21,22,23,24. Here, we show that the reduction of LHFPL3-AS2 in Caco-2 cells substantially affects their spontaneous morphology when grown as a monolayer and their ability to form lumens when grown as 3-dimensional (3D) cysts models. This defect is coupled with mislocalization of the actin cytoskeleton and adherent and tight junction proteins, and altered mitotic spindle formation, which results in a reduction in epithelial proliferation.
Results
LHFPL3-AS2 lncRNA is reduced in CD and in patients with more severe mucosal injury
We previously characterized widespread dysregulation of lncRNAs, including LHFPL3-AS2, in ileal biopsies of the treatment naïve CD pediatric RISK cohort16. We confirmed the reduction of LHFPL3-AS2 expression in bulk mucosal biopsies of CD patients compared to controls in the RISK cohort (Fig. 1A, p = 0.0083) and validated this reduction also in the adult SOURCE cohort (Fig. 1B, p < 0.0001). Interestingly, significant differences in LHFPL3-AS2 expression were not detected in celiac disease, another inflammatory disorder affecting the small intestine (duodenum, SEEM cohort, Supplementary Fig. S1). LHFPL3-AS2 is a relatively uncharacterized lncRNA located on chromosome 7, adjacent and in an antisense orientation to the LHFPL3 protein-coding gene (Supplementary Fig. S1A). While we detected LHFPL3-AS2 expression in the ileum in both RISK and SOURCE cohorts, LHFPL3 located in the same locus, showed minimal or no expression in the ileum (Supplementary Fig. S1C) and is therefore unlikely to be regulated in this context by LHFPL3-AS2. LHFPL3 was also not part of the differentially expressed genes between CD and controls in a recent publication25. Notably, LHFPL3-AS2 was also expressed in isolated primary epithelia in organoids derived from intestinal biopsies from patients undergoing evaluation via endoscopy, and its expression was significantly reduced upon inflammatory triggering with TNFa plus IFNγ, mimicking the reduction seen in the bulk biopsies in CD patients (Fig. 1C). Secondary analysis of a publicly available mRNAseq dataset 26 showed that LHFPL3-AS2 expression in the ileum of healthy human subjects was significantly higher than in the colon (Supplementary Fig. S1D), and this was also true in isolated epithelia in the ileum vs. sigmoid colon27 (Supplementary Fig S1E). The presence of deep ulcers (DUs) in the ileum is known to be associated with endoscopic severity and poorer clinical outcomes28,29,30. Stratifying the RISK CD patients into 2 groups based on the presence or absence of DUs (CD-DU and CD-noDU groups respectively) that was recorded during endoscopy, demonstrated a significant further decrease in LHFPL3-AS2 levels in those with deep ulcers that represent a more severe endoscopic disease (Fig. 1D). These results are further supported by the significant negative correlation in both the RISK and SOURCE cohorts (RISK: r = − 0.5619, SOURCE: r = − 0.8340, p < 0.0001) between levels of the calprotectin encoding gene (S100A8), which is used as a clinical marker of intestinal inflammation, and LHFPL3-AS2 expression (Fig. 1E,F). In contrast, similar to previous analyses that showed that clinical severity does not correlate with ileal gene expression16, 30 and that clinical severity only poorly correlates with endoscopic severity31, no correlation was noted between LHFPL3-AS2 expression and clinical severity (PUCAI).
LHFPL3-AS2 lncRNA is reduced in CD patients and its expression is negatively correlated with CD severity. (A,B) LHFPL3-AS2 mRNA is significantly reduced in bulk mucosal biopsies of CD cases vs. controls in two independent cohorts: the pediatric RISK (A) 213 CD, and 47 controls) and the adult SOURCE (B) 8 CD, and 12 controls) cohorts. (C) LHFPL3-AS2 is expressed in human ileum-derived organoid culture and is reduced upon treatment with 40 ng/ml IFNγ and 20 ng/ml TNFa. (D) LHFPL3-AS2 levels are further reduced in CD patients with deep ulcers (CD-DU, n = 71) in comparison to those without (CD-noDU, n = 78). LHFPL3-AS2 (TPM) values are shown. (E,F) LHFPL3-AS2 log2(TPM) significantly correlates with the calprotectin S100A8 gene as a continuous value in RISK ((E), n = 260) and SOURCE (F, n = 20). (G) Functional annotation enrichment analyses of 470 genes that were co-expressed with LHFPL3-AS2 in RISK using ToppGene/ToppCluster32, 33 and Cytoscape34. GO: Biological Process (orange), GO: Cellular component (green), GO: Molecular function (blue). The full list of functional enrichment results and P values are in Supplementary Dataset S1. A two-sided t-test was calculated between groups and Pearson correlation was used for correlations. *P < 0.05, **P < 0.01, ***P < 0.001.
To gain further insights into the potential functions of LHFPL3-AS2, we analyzed its co-expression within the previously defined core CD signature in RISK16 (2160 protein-coding genes). We identified 470 genes with similar patterns of expression as LHFPL3-AS2 (Pearson correlation, 0.98 < r < 1) across the predefined patient groups (controls, CD-DU, and CD-noDU) with a high likelihood for coregulation and shared biological function. Functional annotation enrichment analyses of these genes using ToppGene32 (Fig. 1G and Supplementary Dataset S1) showed enrichment for terms related to the actin cytoskeleton (FDR P < 1.66E − 07), apical (FDR P < 2.97E − 20) and basal membrane (FDR P < 1.80E − 05), endoplasmic reticulum (ER) membrane (FDR P < 2.33E − 03), transepithelial transport (FDR P < 2.78E − 05), microvillus (FDR P < 3.88E − 15) and intestinal absorption (FDR P < 1.24E − 05). Disease-enriched terms included diarrhea (FDR P < 5.47E − 04) and gastrointestinal diseases (FDR P < 3.17E − 02).
LHFPL3-AS2 knockdown interrupts Caco-2 morphology
Comparing the CD cohorts and Caco-2 mRNAseq analyses indicated that many of the epithelial lncRNAs that were reduced in CD, including LHFPL3-AS2, are expressed in Caco-2 cells16. Caco-2 cells are human-derived colon epithelial cancer cells that can be grown as a polarized monolayer expressing a phenotype resembling that of intestinal enterocytes including tight junctions and metabolizing enzymes35 or as a 3-dimensional (3D) polarized cysts model36. We, therefore, used this cell line as a convenient and robust system to characterize LHFPL3-AS2 and to test the effect of LHFPL3-AS2 loss of function (LOF) on epithelial functions. We first confirmed that LHFPL3-AS2 expression in this system shows a similar pattern as in mucosal biopsies enriched for epithelia using mRNAseq (Supplementary Fig. S2A). Fractionation showed that LHFPL3-AS2 is distributed between the nucleus and the cytoplasm (Supplementary Fig. S2B), and fluorescence RNA in situ hybridization (FISH) showed LHFPL3-AS2 probes localizing to perinuclear and nuclear regions. Since LHFPL3-AS2 cytoplasmic distribution was adjacent to the nucleus, which is the approximate location of the ER, and since in the co-expression analysis of the human cohort we observed an enrichment in ER-related genes, we assessed co-localization of LHFPL3-AS2 with the ER marker calnexin (CANX) and observed substantial overlap (Supplementary Fig. S2D).
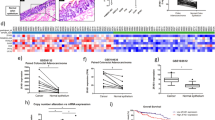
We developed a loss of function (LOF) system to mechanistically model our observations of LHFPL3-AS2 reduction in CD patients. Using CRISPRi and two independent LHFPL3-AS2-specific guide RNAs (g1 and g2 gRNAs, Supplementary Table S2), we depleted LHFPL3-AS2 expression as confirmed by qPCR in comparison to control gRNA (gCtl, Fig. 2A). LHFPL3-AS2 knockdown cells (with either g1 and g2) exhibited abnormal morphology when grown as monolayer (Fig. 2B,C), showing markedly reduced confluency and spreading on the plate, with atypical rounding and clumping that was detected more substantially on the Z-axis (Fig. 2C). This atypical rounding of LHFPL3-AS2 knockdown cells when cells were simply seeded as a monolayer, and the lack of cells confluency hampered our ability, for example, to measure trans-epithelial electrical resistance (TEER) or permeability in the knockdown cells. We, therefore, looked for alternative growth conditions in 3D that will enable head-to-head comparison of LHFPL3-AS2 knockdown and controls. We turned to the Caco-2 cysts 3-dimensional model to systematically test the effects of LHFPL3-AS2 knockdown. In this system, when Caco-2 cells are cultured in Matrigel, they form lumenized 3D cysts36 in which the inside layer is the apical luminal membrane, and the outer membrane represents the basolateral side (Fig. 2D). Bright field imaging (Fig. 2D) and hematoxylin and eosin (H&E, Fig. 2E) staining of the fixed 3D cyst cultures showed that LHFPL3-AS2 knockdown cells failed to form internal luminal cavities. Systematic quantification of cyst morphology indicated that only 10% of LHFPL3-AS2 knockdown cysts formed internal lumens, compared to 62–65% of control cysts (Fig. 2F, p < 0.0001). In addition, F-actin staining showed the expected dominant apical localization in control cysts, whereas in LHFPL3-AS2 knockdown cysts, F-actin was spread throughout the cell membrane (Fig. 2G). We also measured the F-actin intensity signal peaks along the apical-basolateral axis of the 3D cysts as previously described37, which further highlighted the abnormal apical: basolateral distribution of actin in LHFPL3-AS2 knockdown cells (Fig. 2H, Supplementary Fig. S3). Moreover, our microscopic analyses showed internal inclusions of apical markers in LHFPL3-AS2 knockdown cysts Fig. 2F and Supplementary Figs. S3 and S4). This pattern resembles the result of failed protein trafficking, as seen in microvillus inclusion disease, which leads to severe diarrhea and inability to absorb nutrients38.
LHFPL3-AS2 knockdown results in robust loss of Caco-2 luminal cyst formation. (A) LHFPL3-AS2 knockdown was achieved using CRISPRi and 2 gRNAs, and those cells were compared with gCtl. qPCR confirmed a reduction in LHFPL3-AS2, after normalization to GAPDH. The two-sided t-test is shown. ****P < 0.0001. (B) Light microscopy imaging (× 10) of LHFPL3-AS2 knockdown or Caco-2 and gCtl cells after seeding them as a monolayer on a culture plate for 3 days, showing spontaneous rounding and clumping in the knockdown cells. Scalebar—100uM, magnification – × 10. (C) F-actin Phalloidin staining (green) and nuclear staining (blue) of LHFPL3-AS2 knockdown or gCtl cells after seeding them as a monolayer on a culture plate. The upper panel shows the xy-axis views and the lower panel shows a 3D reconstruction of z-stack images. LHFPL3-AS2 knockdown cells show a rounding appearance as demonstrated on the z-axis when grown as a monolayer, with less confluency and more clumping. Magnification x63oil, scalebar-20 µM. (D) Schematic representation and bright field imaging of LHFPL3-AS2 knockdown or Caco-2 and gCtl 3-dimensional (3D) cyst grown in Matrigel for 5 days, with lumen/apical part inside. Scalebar—20uM, magnification – × 40. (E) Haematoxylin and eosin (H&E) staining of Caco-2 cysts shows that most LHFPL3-AS2 knockdown cysts have no lumen. Scalebar—20uM, magnification – × 40. (F) Quantification of (D) indicating the percentage of cysts with a single central lumen, no lumen, or multiple lumens were quantified. Fisher exact test was performed comparing the number of cysts with a single lumen to those with no lumen or with multiple small lumens. (G) Phalloidin staining for F-actin (green) and nuclear Hoechst (blue) staining of Matrigel-grown cysts. Magnification x63oil, scalebar-20uM. (H) Line plots denoting the intensity of actin staining along the apical/basal axis of indicated cyst along the white line, from the apical to basolateral side. The image shows a representative experiment out of 6 replications (shown in Supplementary Fig. S3).
LHFPL3-AS2 knockdown results in a robust reduction in the expression of genes involved in epithelial morphology and structure
We performed mRNASeq analysis of LHFPL3-AS2 knockdown cells and controls that were seeded as monolayers. Principal component analysis (PCA) using all genes that passed expression filtering (n = 11,489) showed a clear separation between the knockdown cells (g1 and g2) and control (Fig. 3A). We identified 584 genes that were differentially expressed between the LHFPL3-AS2 knockdown cells compared to control cells (fold change > = 1.5 and FDR corrected p < = 0.05; Fig. 3B), with the majority of the genes (462/584; 79%) being downregulated upon LHFPL3-AS2 knockdown. Among the reduced genes we found several key regulators of polarity processes, including, for example, RAB17 [fold change (FC) = − 3.2], a GTPase which functions as a regulator of intracellular membrane trafficking primarily in epithelia13, CDC42EP3 (FC = − 3.6), which is involved in actin cytoskeleton re-organization downstream of CDC4239 to induce actin filament assembly leading to cell shape changes, SCRIB (FC = − 6.5), which involved in the establishment of apicobasal cell polarity40, 41, and BMP family genes, which are pivotal for maturation and differentiation of intestinal epithelial cells42. Functional enrichment of the 462 downregulated genes (Fig. 3C and Supplementary Dataset S2) showed enrichment for terms including basal part of the cell (FDR P = 0.00002534) and basal plasma membrane (FDR P = 0.00002534), endoplasmic reticulum compartment (FDR P = 0.02614), transmembrane transport (FDR P = 0.00001377), anchoring junction (FDR P = 0.0006941), regulation of morphogenesis of an epithelium (FDR P 0.00006018, 0.00005167), cell adhesion molecule binding (FDR P = 0.01726), b-catenin-TCFL2 complex (FDR P = 0.02249) and cyclin E1-CDK2 complex (FDR P = 0.02249).
LHFPL3-AS2 knockdown reduced expression of genes, known to regulate epithelial morphology, including CTNNB1. (A) PCA using 11,489 protein-coding genes that passed expression filtering and colored by LHFPL3-AS2 knockdown (g1 and g2 in red) and gCtl (blue). (B) Heatmap of 584 genes that were differentially expressed between LHFPL3-AS2 knockdown cells (g1 and g2) compared to controls (gCtl, fold change > = 1.5 and FDR corrected p < = 0.05). (C) ToppGene32 functional annotation enrichment of 462/584 downregulated gene protein-coding genes with the associated − log10(FDR P value). (D) STRING43 predicted interaction network with visualization using Cytoscape34 of the downregulated genes. (E) qPCR validation of genes that were reduced in the mRNAseq dataset, normalized to GAPDH. The two-sided t-test is shown. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. (F) Schematic representation of Wnt signaling activation by LiCl. (G) Western blots (representative experiment out of 3 replications) of LHFPL3-AS2 knockdown cells (g1 and g2) and controls (gCtl) with and without LiCl to activate the Wnt pathway showing reduction of CTNNB1 and TCF4 at baseline and after LiCl in LHFPL3-AS2 knockdown cells, and of CDH1 with LiCl.
Using String, a functional protein association networks tool, we connected some of the downregulated genes to β-catenin (CTNNB1), which was identified as a central node in this sub-network (Fig. 3D). We validated the reduced expression of CTNNB1, TCF4, WT1, CDH11, SCRIB, TFRC, and MYO1D in LHFPL3-AS2 knockdown cells by qPCR (Fig. 3E), and the reduction in CTNNB1 and TCF4 at the protein level using western blotting (Fig. 3G). CTNNB1 and TCF4 are part of the canonical Wnt signaling pathway, which plays a role in the development and renewal of the intestinal epithelium; WT1 and MYO1D are involved in cell morphology while CDH11 and SCRIB are part of the adhesion process. Next, we stimulated Caco-2 cells with Lithium chloride (LiCl), which is known to activate Wnt/β-catenin signaling by inhibiting the GSK3 activity37, 44, thereby increasing the free β-catenin fraction that translocates to the nucleus to activate the TCF4 transcription factor and its downstream genes (Fig. 3F scheme). A similar reduction was noted in TCF4 and CTNNB1 levels in LHFPL3-AS2 knockdown cells at baseline and after LiCl treatment, but we also documented a reduction in E-cadherin (CDH1) more persistently upon LiCl treatment. CDH1 is a known adherens junction protein and a downstream target of CTNNB1 (Fig. 3G)45, 46. We next examined the distribution of tight and adherens junction proteins by immunofluorescence in Caco-2 3D cysts. Staining for these markers showed that in knockdown cells, most were abnormally distributed (Supplementary Fig. S4). Specifically, the apical markers F-actin and ZO-1 were expressed in a non-homogenous pattern with intracellular vesical-like structures in cysts that did not form lumens, and the junction marker CDH1 similarly localized intracellularly in clusters in LHFPL3-AS2 downregulated cells (Supplementary Fig. S4A). We also captured reduced ZO-1 and E-cadherin CDH1 mRNA levels in LHFPL3-AS2 downregulated cells (Supplementary Fig. S4B-C). Staining for the intercellular junctional markers CTNNB1 and JAM-A also showed disorganized phenotypes in LHFPL3-AS2 down-regulated cysts (Supplementary Fig. S4D).
LHFPL3-AS2 downregulation induces abnormal mitotic divisions with multipolar spindles, and reduced proliferation
RNAseq analyses of LHFPL3-AS2 knockdown cells further indicated significant downregulation of cyclin E1 (CCNE1) and CDK2, a complex that is required for cell cycle G1/S transition (Supplementary Dataset S2 and Fig. 4A), and we validated the reduction in CCNE1 and CDK2 at the protein level (Fig. 4B). This observation, together with the known role of CTNNB1 and Wnt in regulating proliferation, prompted us to explore whether LHFPL3-AS2 affects cell division. Indeed, LHFPL3-AS2 knockdown resulted in an increase in the cell fraction arrested at the G1 phase, with subsequent reduction in G2/M (Fig. 4C and Supplementary Fig. S5 showing representative FACS results). The G1 arrest in LHFPL3-AS2 knockdown cells led to attenuated cell proliferation as measured by decreased colony formation (Fig. 4D), and XTT assay (Fig. 4E) in monolayer cells, as well as Ki67 staining, a nuclear protein that is associated with cellular proliferation, in cysts (Fig. 4F).
LHFPL3-AS2 downregulation inhibited cell proliferation resulting in G1 cell cycle arrest. (A) mRNA levels (TPM) of CCNE1 and CDK2 in LHFPL3-AS2 knockdown cells (g1 and g2) and controls (gCtl). (B) Western Blot analysis of CCNE1 and CDK2 using GAPDH as a loading control (representative experiment out of 3 replications). Quantifying the protein expression, using ImageJ, and normalizing it to GAPDH indicated a reduction in CCNE1 of 59% with g1 and 24% with g2 and a more modest reduction in CDK2 of 16% with g1 and 18% with g2. (C) Propidium Iodide staining (PI) and FACS analyses of LHFPL3-AS2 knockdown cells (g1 and g2) and controls (gCtl). Percentages of cells in G1, S, and G2/M cell-cycle phases are in a pie chart. One sided t-test, n = 4, *p < 0.05, **p < 0.01. (D) Colony formation assay with representative images (left panel) and quantification (right panel) of the colonies (n = 3). (E) XTT assay absorbance values (OD) (n = 3) show that suppression of LHFPL3-AS2 inhibited Caco-2 cell proliferation. (F) Immunohistochemistry (IHC) staining of Ki-67 in paraffin-embedded sections of Caco-2 3D cysts. Scalebar – 10uM, magnification – × 40.
Cell cycle arrest has been linked with several centrosome defects, such as centriole loss, separation, and fragmentation47. This together with the effect of LHFPL3-AS2 reduction on cyst lumen formation, epithelial polarity, and cell proliferation suggested that LHFPL3-AS2 loss may affect a mitotic and cell division rearrangement. By staining for α-tubulin and DNA (Hoechst), we were able to capture several mitotic events in Caco-2 cysts. Whereas in the control cysts we observed the normal bipolar mitotic spindle, in LHFPL3-AS2 knockdown cysts we noted a disorganized multipolar pattern (Fig. 5A and B). Aiming to capture more mitotic events, we used Taxol (Paclitaxel, 20 nM for 24 h), which stabilizes microtubules and reduces their dynamics in the G2/M phase, resulting in mitotic arrest48. Upon Taxol treatment, LHFPL3-AS2 knockdown resulted in a significant increase in multipolar spindle formation and centrosome amplification (Fig. 5C, white arrows, Supplementary Fig. S6). Indeed, 80–81% of cells in LHFPL3-AS2 knockdown cysts showed multipolar spindles compared to just 11% of control cells (Fig. 5C, Supplementary Videos 1–3).
LHFPL3-AS2 knockdown results in abnormal multipolar mitotic spindle formation. (A) Fluorescence staining using anti-tubulin (red) and nuclear Hoechst (blue) of LHFPL3-AS2 CRISPRi knockdown and gCtl Caco-2 cysts. White arrows indicate mitotic spindles. Scalebar—20uM, magnification—x60oil. (B) Schematic representation of a normal bipolar mitotic spindle. (C) Caco-2 grown as 3D cysts were treated with 20 nM Taxol for 24 h to arrest cells during mitosis. Left panel—representative images of the mitotic spindles in LHFPL3-AS2 knockdowns and controls (gCtl). White arrows indicate centrosomes. Right panel—Percentage of cells with bipolar spindles, multipolar spindles, or monopolar spindles captured during mitosis. Fisher exact test between the fraction of cells with bi-polar spindles vs. multi/mono-polar spindles. ****P ≤ 0.0001. (D) Schematic cartoon summarizing LHFPL3-AS2's potential role in regulating epithelial polarity, proliferation, and mitotic spindle formation. Reduction of LHFPL3-AS2 affects apicobasal positioning of cytoskeleton protein (actin) and tight/adherens junction proteins. Loss of cellular polarity and abnormal cellular positioning likely underlies the abnormal mitotic spindle and centrosome arrangement, resulting in G1 cell cycle arrest, reduced cellular proliferation, and the robust subsequent inhibition of transcription and translation of many key epithelial genes including CTNNB1 and TCF4.
Discussion
The apical and basal plasma membrane domains are differentially equipped with proteins to control the absorption and secretion of nutrients between the gut lumen, cell interior, and the intestine. Defects in apicobasal polarity disrupt the epithelial barrier defense against pathogens and affect cellular metabolism and uptake of nutrients49, and disruption in these epithelial functions contributes to CD pathogenesis3. Herein, we highlight a significant reduction of LHFPL3-AS216, a novel, relatively uncharacterized lncRNA, in two independent CD datasets (RISK, SOURCE). In addition, we show that LHFPL3-AS2 is further downregulated in patients with more severe disease, as determined by the presence of deep ulcers, and with higher mucosal calprotectin, a marker of intestinal inflammation assessment used in clinic, mRNA levels. To identify potential LHFPL3-AS2 functions, we performed co-expression analysis ("guilt-by-association") in the human cohort50, and showed enrichment for genes linked with the actin cytoskeleton, apical and basal membranes, and the ER sub-compartment. We demonstrated that LHFPL3-AS2 is predominantly expressed in the small rather than large intestine, is distributed between the nucleus and cytoplasm, and colocalizes with the ER marker Calnexin in Caco-2 cells.
Depletion of LHFPL3-AS2 in Caco-2 cells resulted in a robust spontaneous alteration of cellular morphology when cells were grown as a monolayer. LHFPL3-AS2-depleted cells rounded up and clumped and for that reason, we could not reach cellular confluency and were unable to measure for example TEER or permeability. Since LHFPL3-AS2 knockdown demonstrated a more 3D structure, it encouraged us to turn to the more challenging 3D cyst-polarized Caco-2 model for imaging and quantification of the cellular abnormalities of both the knockdowns and the controls in the same conditions. Using the 3D cyst-polarized model we were able to show substantial and significant defective lumen formation and altered actin distribution at the apicobasal axis. Cell rounding and insufficient spreading may interpret as diminished cell-extracellular matrix interactions, but the defects seen in the lumen formation in 3D, more likely suggest an abnormal polarity. Respectively, we observed that LHFPL3-AS2-depleted cysts retained internal inclusions of apical markers (i.e., actin). This intracellular vesicular pattern suggests a failure of protein trafficking, rather than pure differentiation defects, which is also observed in polarity-associated disorders like microvillus inclusion disease (MVID) that leads to severe diarrhea and inability to absorb nutrients in the intestine. Diarrhea and poor absorption are also key components of CD pathogenesis. Moreover, proper epithelial renewal and proliferation is part of the mucosal healing process, which is the main treatment goal in CD51, and our results suggest that this process is also attenuated by LHFPL3-AS2 knockdown, which can lead to abnormal healing.
Differential RNAseq expression analysis further supported this concept. We identified a reduction of 462 genes in LHFPL3-AS2-depleted cells including several that are known to have key roles in cellular polarity, such as RAB17, an epithelial cell-specific GTPase13, and CDC42EP3, which acts downstream of CDC4239 to induce actin filament assembly leading to cell shape changes, BMP family proteins which affect cell differentiation42, and SCRIB which regulates the establishment of apicobasal cell polarity40, 41 and the progression from G1-S cell cycle phase41, 52. Functional enrichment of the reduced genes highlighted pathways linked with the apical cell membrane, actin filament bundle, microvillus, regulation of morphogenesis of epithelium, adherens junctions, β-catenin-TCF4 complex, and connection to the ER compartment. Membrane proteins are synthesized in the ER and transferred to the trans-Golgi network, where they are sorted into distinct endosomal carrier vesicles and transported to the apical or basolateral plasma membrane53. Failure of vesicle trafficking causes apical proteins to accumulate sub-apically and can result in microvillus inclusion disease14. Previous studies have indicated that assembly of the E-cadherin/β-catenin complex occurs in the very early stage of the intracellular trafficking54,55,56, and that this process is the obligatory step in efficient export from the ER and for subsequent delivery of E-cadherin to the basal-lateral membrane of polarized MDCK cells56. We identified CTNNB1 as a central node in the network of reduced genes resulting from LHFPL3-AS2 knockdown. The overall effect observed from the reduction of LHFPL3-AS2 knockdown is summarized in Fig. 5D; the reduction of LHFPL3-AS2 affects epithelial polarity and apicobasal positioning of cytoskeleton protein (actin) and tight/adherens junction proteins. Loss of cellular polarity and abnormal cellular positioning likely underlies the abnormal mitotic spindle and centrosome arrangement, resulting in G1 cell cycle arrest, reduced proliferation, and subsequent inhibition of transcription and translation of many key epithelial genes including CTNNB1 and TCF4. LHFPL3-AS2 may participate in regulating intracellular trafficking and epithelial polarity through its cytoplasmic ER localization, but further studies are needed to support this idea.
Cell division provides a mechanism for maintaining the structural integrity of the epithelial barrier in rapidly proliferating tissues, such as the intestine57. Notably, centrosome duplication, the initial event in cell division, takes place during G1–S transition and defects in centrosome positioning can disrupt epithelial homeostasis58. LHFPL3-AS2 silencing resulted in reduction in CCNE1 and CDK2, which regulate the transition from G1 to S phase59. We also noted that LHFPL3-AS2 knockdown cysts displayed multipolar spindle formation and abnormal centrosome integrity. Similar to the effects seen here, ZO-1-deficient colonic epithelia have aberrantly oriented mitotic spindles, resulting in abortive proliferation60. Loss of centrosome integrity was recently shown to trigger checkpoint regulators that inhibit G1–S progression47, and cells arrested in the G1 phase can assemble more than four centrioles61. Other lncRNAs like gadd7, which destabilizes CDK6 mRNA21, were previously shown to control cell cycle progression via modulation of the expression of critical cell cycles regulators such as cyclins, CDKs, and CDK inhibitors20,21,22,23,24, but here we anticipate inhibition of proliferation and the G1 arrest may be secondary to improper epithelial polarity linked with abnormal spindle positioning. LHFPL3-AS2 is a relatively uncharacterized lncRNA, with one recent publication indicating that it suppresses metastasis of non-small cell lung cancer (NSCLC) by interacting with SFPQ to regulate TXNIP expression62. Our RNA-seq analyses however identified TXNIP to be up-regulated upon LHFPL3-AS2 reduction. Importantly, over-expression of TXNIP (as seen here as a result of LHFPL3-AS2 reduction in Caco-2 cells) is known to induce G0/G1 cell cycle arrest63 which is consistent with our observation. A possible explanation for discrepancies between these studies may be that lncRNAs have different expression profiles and mechanisms of action in different cell types. For instance, HNF1A-AS1 lncRNA has been defined as a tumor suppressor gene in laryngeal squamous cell carcinoma64, but also has an oncogenic role in gastric cancer65. It is also possible that regulatory RNAs such as miRNA and circRNA mediate the effect seen upon LHFPL3-AS2 reduction, and this concept will need to be further explored.
A notable strength of our study was the use of treatment-naïve adult and pediatric patient cohorts, free from confounding variables of previous therapy. We used diverse, complementary wet-lab and mRNAseq informatics approaches to cross-validate our results, which altogether support a model whereby LHFPL3-AS2 regulates intestinal epithelial functions. However, our use of treatment-naïve samples is also a limitation, as the associated inflammation may confound results. Additionally, for studying the effect of LHFPL3-AS2 knockdown, we used a tissue culture model rather than an animal model or organoid system. However, human-derived cell lines are widely used in recently published work to understand the role of long non-coding RNAs (lncRNAs) in human-related systems66,67 and functions. Although Caco-2 cells have hyper-tetraploid karyotypes, we showed that in most of the cases (89%) their mitotic spindles had a normal bipolar phenotype, unlike the LHFPL3-AS2 3D cysts. Furthermore, we confirmed that Caco-2 cells show a similar pattern of LHFPL3-AS2 expression as that seen in mucosal biopsies enriched for epithelia, indicating that it is an appropriate model system. Finally, while we saw a robust effect on spontaneous cellular morphology on cells grown as monolayer and in 3D polarized cyst culture when LHFPL3-AS2 was inhibited by 2 independent gRNAs, we did not fully explore the mechanism by which LHFPL3-AS2 regulates those functions. However, RNAseq data of the knockdown cells did show a significant and substantial reduction in polarity-associated genes (SCRIB was reduced with a fold change (FC) = − 6.5, CDC42EP3 FC = − 3.6, and RAB17 with FC = − 3.2). LHFPL3-AS2 rescue experiments are valuable to confirm LHFPL3-AS2 role but will be part of future work. It will also be interesting to examine if CTNNB1 over-expression, which is a predicated candidate of the enrichment analyses of the LHFPL3-AS2 knockdown, is able to rescue the LHFPL3-AS2 phenotypes.
In conclusion, we show that LHFPL3-AS2 controls epithelial morphogenesis by regulating epithelial polarity, mitotic spindle formation, and proliferation. Epithelial polarity and proliferation are key in maintaining epithelial homeostasis and functions that are relevant to CD. Reduced expression of LHFPL3-AS2 in CD and its further reduction in more severe CD forms emphasizes the clinical significance in this context. Future studies are needed to determine how LHFPL3-AS2 regulates epithelial polarity potentially via its localization to the ER.
Materials and methods (see supplementary methods)
Cohorts, study approval, and RNAseq analyses
We performed secondary analyses of the published pediatric RISK (GSE101794) and the adult SOURCE (GSE199906) treatment naïve CD cohorts to test the baseline expression of LHFPL3-AS2 in CD in comparison to controls in the ileum. Secondary analyses of other published cohorts were used for expression validation of LHFPL3-AS2 in the ileum and colon tissue in healthy controls in bulk biopsies26, and in isolated epithelia26. Sheba Medical Center Institutional Review Board approved the SOURCE CD cohort (Supplementary Table S1) protocol and informed consent was obtained. LHFPL3-AS2 expression was assessed after uniformly reanalyzing the transcriptomics raw FATSQ files. RNA-seq on LHFPL3-AS2 knockdown Caco-2 cells was performed using Lexogen QuantSeq 3' mRNA-Seq libraries sequencing (data was deposited in GEO: GSE216810). Principal Coordinates Analysis (PCA) was performed to summarize differences between knockdown and control conditions. In downstream analysis, 11,489 protein-coding mRNA genes with Reads per Milion (RPM) above 3 in 20% of the samples were included. Differentially expressed genes were determined in GeneSpring® software with fold change differences (FC) ≥ 1.5 and using the Benjamini–Hochberg false discovery rate correction (FDR ≤ 0.05). ToppGene32, ToppCluster software were used to perform functional annotation enrichments, and Cytoscape.v3.0.268 was used for visualization.
LHFPL3-AS2 knockdown in cell culture
LHFPL3-AS2 stable knockdown was achieved by infecting Caco-2 cells with CRISPR/dCAS9-KRAB inactivator plasmid that was packaged into virions generated in HEK293T cells using a lentiviral vector system. Monoclonal clones were selected with Puromycin. Then, the pKLV-gRNAs clones which contained the U6 promoter and hygromycin gene (hygro) were packaged into virions and generated in HEK293T cells using a lentiviral vector system. Afterward, a stable clone of Caco-2 expressing CRISPRi plasmid was infected by a lentivirus carrying the specific gRNA or by an empty plasmid that is used as a control (gCtl). gRNA sequences and qPCR primers are in Supplementary Tables S2 and S3 respectively.
Confocal immunofluorescence
Cells were grown as monolayers on coverslips in 12 well plates, or Matrigel in 24 well plates for 3D cysts, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 and blocked in 5% BSA. Then, the cells were incubated with primary antibodies diluted in 5% BSA overnight at 4C, the coverslips were fluorescently labeled and incubated with a secondary antibody (anti-rabbit or anti-mouse Cy3 or Cy5; Abcam). For visualization of F-actin, Phalloidin (in 5% BSA; Abcam) staining was performed for 20 min, followed by Hoescht staining (Sigma). The coverslips were examined using Zeiss confocal microscope (Carl Zeiss AB, Stockholm, Sweden). Images were processed using ZEN 3.1 (blue edition) browser software. For the 3D system, images were produced using a Z-stack confocal-based scan of the cysts. Movies were produced from Z-stack images using the ZEN 3.1 (blue edition) browser. For mitotic spindle visualization, Caco-2 3D cysts were treated with 20 nM Taxol for 24 h and stained with a-Tubulin. Mono/bi/multi-polar spindles were quantified manually, across the Z-stack of the cyst. Apical-basal actin intensity was determined, using the profile feature, within the ZEN 3.1 software, which shows intensity to distance curve of the region of interest (luminal apical to the basolateral side of the cyst). For RNA fluorescent in situ hybridization (FISH), Caco-2 cells were seeded on 18 mm glass coverslips in a 12-well plate. Stellaris RNA FISH was used to visualize LHFPL3-AS2 and CANX antibody was used for co-localization with ER, the cells were fixed and stained according to Stellaris protocol (Biosearch Technologies). Confocal imaging was performed with a Zeiss confocal microscope using a 63 × oil immersion objective.
Proliferation assays
For colony formation assay, Caco-2 cells were seeded (1,000 cells/6 well plate) and cultured for 13 days to form colonies, fixed with 100% ethanol, and stained with 10% Giemsa, photographed, and counted. For cell viability assay, Caco-2 cells were seeded into a 96-well plate, and proliferation was assessed using the XTT Cell Proliferation Assay Kit (Biological Industries) following the manufacturer's protocols. For proliferation assessment in 3D cysts, paraffin-embedded sections of the cysts were stained with Ki67 antibody.
Fluorescence-activated cell sorting (FACS) cell cycle analyses
Cells were fixed in 70% ice-cold ethanol at 4 °C and stained with 5 mg/ml propidium iodide (Sigma) Fluorescence intensity was analyzed using Navios flow cytometer (Beckman Coulter). The analysis was performed using Kaluza software.
Ethical considerations
The published pediatric RISK (GSE101794) and adult SOURCE (GSE199906) treatment naïve CD cohorts were used to test the baseline expression of LHFPL3-AS2 in CD ileum. Other published cohorts were used for expression validation of LHFPL3-AS2 in the ileum and colon tissue in healthy controls in bulk biopsies26, and in isolated epithelia26. Sheba Medical Center Institutional Review Board approved the SOURCE CD cohort protocol and informed consent was obtained.
Data availability
mRNAseq generated as part of this study were deposited in GEO: GSE216810. Other datasets used include the RISK (GSE101794) and the SOURCE (GSE199906) cohorts. All authors had access to all data and reviewed and approved the final manuscript.
References
Magie, C. R. & Martindale, M. Q. Cell-cell adhesion in the cnidaria: Insights into the evolution of tissue morphogenesis. Biol. Bull. 214, 218–232. https://doi.org/10.2307/25470665 (2008).
Rodriguez-Boulan, E. & Macara, I. G. Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15, 225–242. https://doi.org/10.1038/nrm3775 (2014).
Guo, C. & Shen, J. Cytoskeletal organization and cell polarity in the pathogenesis of Crohn’s disease. Clin. Rev. Allergy Immunol. 60, 164–174. https://doi.org/10.1007/s12016-020-08795-5 (2021).
Jewett, C. E. & Prekeris, R. Insane in the apical membrane: Trafficking events mediating apicobasal epithelial polarity during tube morphogenesis. Traffic https://doi.org/10.1111/tra.12579 (2018).
van der Wouden, J. M., Maier, O., van IJzendoorn, S. C. & Hoekstra, D. Membrane dynamics and the regulation of epithelial cell polarity. Int. Rev. Cytol. 226, 127–164. https://doi.org/10.1016/s0074-7696(03)01003-9 (2003).
Weisz, O. A. & Rodriguez-Boulan, E. Apical trafficking in epithelial cells: Signals, clusters and motors. J. Cell Sci. 122, 4253–4266. https://doi.org/10.1242/jcs.032615 (2009).
Klunder, L. J., Faber, K. N., Dijkstra, G. & van IJzendoorn, S. C. Mechanisms of cell polarity-controlled epithelial homeostasis and immunity in the intestine. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a027888 (2017).
Akhmanova, A. & Hoogenraad, C. C. Microtubule minus-end-targeting proteins. Curr. Biol. 25, R162-171. https://doi.org/10.1016/j.cub.2014.12.027 (2015).
Bartolini, F. & Gundersen, G. G. Generation of noncentrosomal microtubule arrays. J. Cell Sci. 119, 4155–4163. https://doi.org/10.1242/jcs.03227 (2006).
Dammermann, A., Desai, A. & Oegema, K. The minus end in sight. Curr. Biol. 13, R614-624. https://doi.org/10.1016/s0960-9822(03)00530-x (2003).
Oceguera-Yanez, F. et al. Ect2 and MgcRacGAP regulate the activation and function of Cdc42 in mitosis. J. Cell Biol. 168, 221–232. https://doi.org/10.1083/jcb.200408085 (2005).
Hirsch, D. S., Pirone, D. M. & Burbelo, P. D. A new family of Cdc42 effector proteins, CEPs, function in fibroblast and epithelial cell shape changes. J. Biol. Chem. 276, 875–883. https://doi.org/10.1074/jbc.M007039200 (2001).
Zacchi, P. et al. Rab17 regulates membrane trafficking through apical recycling endosomes in polarized epithelial cells. J. Cell Biol. 140, 1039–1053. https://doi.org/10.1083/jcb.140.5.1039 (1998).
Ameen, N. A. & Salas, P. J. Microvillus inclusion disease: A genetic defect affecting apical membrane protein traffic in intestinal epithelium. Traffic 1, 76–83. https://doi.org/10.1034/j.1600-0854.2000.010111.x (2000).
Odenwald, M. A. & Turner, J. R. The intestinal epithelial barrier: A therapeutic target?. Nat. Rev. Gastroenterol. Hepatol. 14, 9–21. https://doi.org/10.1038/nrgastro.2016.169 (2017).
Haberman, Y. et al. Long ncRNA landscape in the ileum of treatment-naive early-onset Crohn disease. Inflamm. Bowel Dis. 24, 346–360. https://doi.org/10.1093/ibd/izx013 (2018).
Lee, H., Zhang, Z. & Krause, H. M. Long noncoding RNAs and repetitive elements: Junk or intimate evolutionary partners?. Trends Genet. 35, 892–902. https://doi.org/10.1016/j.tig.2019.09.006 (2019).
Xiao, L. et al. Long noncoding RNA SPRY4-IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Mol. Biol. Cell 27, 617–626. https://doi.org/10.1091/mbc.E15-10-0703 (2016).
Zou, T. et al. H19 long noncoding RNA regulates intestinal epithelial barrier function via MicroRNA 675 by interacting with RNA-binding protein HuR. Mol. Cell. Biol. 36, 1332–1341. https://doi.org/10.1128/MCB.01030-15 (2016).
Kotake, Y. et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30, 1956–1962. https://doi.org/10.1038/onc.2010.568 (2011).
Liu, X., Li, D., Zhang, W., Guo, M. & Zhan, Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 31, 4415–4427. https://doi.org/10.1038/emboj.2012.292 (2012).
Tripathi, V. et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 9, e1003368. https://doi.org/10.1371/journal.pgen.1003368 (2013).
Wang, X. et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454, 126–130. https://doi.org/10.1038/nature06992 (2008).
Kitagawa, M., Kitagawa, K., Kotake, Y., Niida, H. & Ohhata, T. Cell cycle regulation by long non-coding RNAs. Cell. Mol. Life Sci. 70, 4785–4794. https://doi.org/10.1007/s00018-013-1423-0 (2013).
Braun, T. et al. Mucosal transcriptomics highlight lncRNAs implicated in ulcerative colitis, Crohn’s disease, and celiac disease. JCI Insight https://doi.org/10.1172/jci.insight.170181 (2023).
Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406. https://doi.org/10.1074/mcp.M113.035600 (2014).
Howell, K. J. et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology 154, 585–598. https://doi.org/10.1053/j.gastro.2017.10.007 (2018).
Allez, M. et al. Long term outcome of patients with active Crohn’s disease exhibiting extensive and deep ulcerations at colonoscopy. Am. J. Gastroenterol. 97, 947–953. https://doi.org/10.1111/j.1572-0241.2002.05614.x (2002).
Mary, J. Y. & Modigliani, R. Development and validation of an endoscopic index of the severity for Crohn’s disease: A prospective multicentre study. Groupe d’Etudes Therapeutiques des Affections Inflammatoires du Tube Digestif (GETAID). Gut 30, 983–989. https://doi.org/10.1136/gut.30.7.983 (1989).
Haberman, Y. et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest. 124, 3617–3633. https://doi.org/10.1172/JCI75436 (2014).
Turner, D. et al. Which PCDAI version best reflects intestinal inflammation in pediatric Crohn disease?. J. Pediatr. Gastroenterol. Nutr. 64, 254–260. https://doi.org/10.1097/MPG.0000000000001227 (2017).
Chen, J., Bardes, E. E., Aronow, B. J. & Jegga, A. G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305-311. https://doi.org/10.1093/nar/gkp427 (2009).
Kaimal, V., Bardes, E. E., Tabar, S. C., Jegga, A. G. & Aronow, B. J. ToppCluster: A multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 38, W96-102. https://doi.org/10.1093/nar/gkq418 (2010).
Saito, R. et al. A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076. https://doi.org/10.1038/nmeth.2212 (2012).
Hidalgo, I. J., Raub, T. J. & Borchardt, R. T. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 96, 736–749 (1989).
Jaffe, A. B., Kaji, N., Durgan, J. & Hall, A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J. Cell Biol. 183, 625–633. https://doi.org/10.1083/jcb.200807121 (2008).
Durgan, J., Kaji, N., Jin, D. & Hall, A. Par6B and atypical PKC regulate mitotic spindle orientation during epithelial morphogenesis. J. Biol. Chem. 286, 12461–12474. https://doi.org/10.1074/jbc.M110.174235 (2011).
Knowles, B. C. et al. Myosin Vb uncoupling from RAB8A and RAB11A elicits microvillus inclusion disease. J. Clin. Invest. 124, 2947–2962. https://doi.org/10.1172/JCI71651 (2014).
Melendez, J. et al. Cdc42 coordinates proliferation, polarity, migration, and differentiation of small intestinal epithelial cells in mice. Gastroenterology 145, 808–819. https://doi.org/10.1053/j.gastro.2013.06.021 (2013).
Qin, Y., Capaldo, C., Gumbiner, B. M. & Macara, I. G. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J. Cell Biol. 171, 1061–1071. https://doi.org/10.1083/jcb.200506094 (2005).
Zhan, L. et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 135, 865–878. https://doi.org/10.1016/j.cell.2008.09.045 (2008).
Zhang, Y. & Que, J. BMP signaling in development, stem cells, and diseases of the gastrointestinal tract. Annu. Rev. Physiol. 82, 251–273. https://doi.org/10.1146/annurev-physiol-021119-034500 (2020).
Szklarczyk, D. et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. https://doi.org/10.1093/nar/gkaa1074 (2021).
Vidal, F. et al. Lithium reduces tumorigenic potential in response to EGF signaling in human colorectal cancer cells. Int. J. Oncol. 38, 1365–1373. https://doi.org/10.3892/ijo.2011.955 (2011).
Gottardi, C. J., Wong, E. & Gumbiner, B. M. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J. Cell Biol. 153, 1049–1060. https://doi.org/10.1083/jcb.153.5.1049 (2001).
Stockinger, A., Eger, A., Wolf, J., Beug, H. & Foisner, R. E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J. Cell Biol. 154, 1185–1196. https://doi.org/10.1083/jcb.200104036 (2001).
Mikule, K. et al. Loss of centrosome integrity induces p38–p53-p21-dependent G1-S arrest. Nat. Cell Biol. 9, 160–170. https://doi.org/10.1038/ncb1529 (2007).
Tishler, R. B., Schiff, P. B., Geard, C. R. & Hall, E. J. Taxol: A novel radiation sensitizer. Int. J. Radiat. Oncol. Biol. Phys. 22, 613–617. https://doi.org/10.1016/0360-3016(92)90888-o (1992).
Danielsen, E. M. & Hansen, G. H. Lipid raft organization and function in the small intestinal brush border. J. Physiol. Biochem. 64, 377–382. https://doi.org/10.1007/BF03174093 (2008).
Sosnovski, K. E. et al. GATA6-AS1 regulates intestinal epithelial mitochondrial functions, and its reduced expression is linked to intestinal inflammation and less favorable disease course in ulcerative colitis (UC). J. Crohns Colitis https://doi.org/10.1093/ecco-jcc/jjad006 (2023).
Froslie, K. F., Jahnsen, J., Moum, B. A., Vatn, M. H. & Group, I. Mucosal healing in inflammatory bowel disease: Results from a Norwegian population-based cohort. Gastroenterology 133, 412–422. https://doi.org/10.1053/j.gastro.2007.05.051 (2007).
Nagasaka, K. et al. Human homolog of Drosophila tumor suppressor Scribble negatively regulates cell-cycle progression from G1 to S phase by localizing at the basolateral membrane in epithelial cells. Cancer Sci. 97, 1217–1225. https://doi.org/10.1111/j.1349-7006.2006.00315.x (2006).
Massey-Harroche, D. Epithelial cell polarity as reflected in enterocytes. Microsc. Res. Tech. 49, 353–362. https://doi.org/10.1002/(SICI)1097-0029(20000515)49:4%3c353::AID-JEMT4%3e3.0.CO;2-8 (2000).
Ozawa, M. & Kemler, R. Molecular organization of the uvomorulin-catenin complex. J. Cell Biol. 116, 989–996. https://doi.org/10.1083/jcb.116.4.989 (1992).
Hinck, L., Nathke, I. S., Papkoff, J. & Nelson, W. J. Dynamics of cadherin/catenin complex formation: novel protein interactions and pathways of complex assembly. J. Cell. Biol. 125, 1327–1340. https://doi.org/10.1083/jcb.125.6.1327 (1994).
Chen, Y. T., Stewart, D. B. & Nelson, W. J. Coupling assembly of the E-cadherin/beta-catenin complex to efficient endoplasmic reticulum exit and basal-lateral membrane targeting of E-cadherin in polarized MDCK cells. J. Cell Biol. 144, 687–699. https://doi.org/10.1083/jcb.144.4.687 (1999).
Reinsch, S. & Karsenti, E. Orientation of spindle axis and distribution of plasma membrane proteins during cell division in polarized MDCKII cells. J. Cell Biol. 126, 1509–1526. https://doi.org/10.1083/jcb.126.6.1509 (1994).
Hebert, A. M., DuBoff, B., Casaletto, J. B., Gladden, A. B. & McClatchey, A. I. Merlin/ERM proteins establish cortical asymmetry and centrosome position. Genes Dev. 26, 2709–2723. https://doi.org/10.1101/gad.194027.112 (2012).
Lingle, W. L., Lukasiewicz, K. & Salisbury, J. L. Deregulation of the centrosome cycle and the origin of chromosomal instability in cancer. Adv. Exp. Med. Biol. 570, 393–421. https://doi.org/10.1007/1-4020-3764-3_14 (2005).
Kuo, W. T. et al. The tight junction protein ZO-1 is dispensable for barrier function but critical for effective mucosal repair. Gastroenterology 161, 1924–1939. https://doi.org/10.1053/j.gastro.2021.08.047 (2021).
Durcan, T. M. et al. Centrosome duplication proceeds during mimosine-induced G1 cell cycle arrest. J. Cell. Physiol. 215, 182–191. https://doi.org/10.1002/jcp.21298 (2008).
Cheng, Z. et al. Long noncoding RNA LHFPL3-AS2 suppresses metastasis of non-small cell lung cancer by interacting with SFPQ to regulate TXNIP expression. Cancer Lett. 531, 1–13. https://doi.org/10.1016/j.canlet.2022.01.031 (2022).
Yamaguchi, F. et al. Rare sugar D-allose induces specific up-regulation of TXNIP and subsequent G1 cell cycle arrest in hepatocellular carcinoma cells by stabilization of p27kip1. Int. J. Oncol. 32, 377–385 (2008).
Shi, Y. et al. Aberrant methylationmediated decrease of lncRNA HNF1AAS1 contributes to malignant progression of laryngeal squamous cell carcinoma via EMT. Oncol. Rep. 44, 2503–2516. https://doi.org/10.3892/or.2020.7823 (2020).
Liu, H. T., Liu, S., Liu, L., Ma, R. R. & Gao, P. EGR1-mediated transcription of lncRNA-HNF1A-AS1 promotes cell-cycle progression in gastric cancer. Cancer Res. 78, 5877–5890. https://doi.org/10.1158/0008-5472.CAN-18-1011 (2018).
Wong, L. S. et al. In vivo genome-wide CRISPR activation screening identifies functionally important long noncoding RNAs in hepatocellular carcinoma. Cell. Mol. Gastroenterol. Hepatol. 14, 1053–1076. https://doi.org/10.1016/j.jcmgh.2022.07.017 (2022).
Sondergaard, J. N. et al. CCT3-LINC00326 axis regulates hepatocarcinogenic lipid metabolism. Gut 71, 2081–2092. https://doi.org/10.1136/gutjnl-2021-325109 (2022).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. https://doi.org/10.1101/gr.1239303 (2003).
Acknowledgements
We gratefully acknowledge RISK and SOURCE co-investigators. We also thank all participants in the study.
Funding
This work was supported by the ERC starting grant (YH, grant No 758313), the Israel Science Foundation (YH, grant No 908/15, 785/22), the I-CORE program (YH, grants No. 41/11), the Helmsley Charitable Trust, and NIDDK P30 DK078392 (Integrative Morphology and Gene Expression Cores). The funding sources did not play a role in the writing of the manuscript or the decision to submit it for publication and did not play a role in data collection, analysis, interpretation; trial design; patient recruitment; or any aspect of the study.
Author information
Authors and Affiliations
Contributions
K.E.S., L.A.D., and Y.H. conceived and designed the study, analyzed the data and wrote the first draft of the manuscript. T.B., A.A., M.B.S., H.A.E., R.B.S., L.A., C.A., and I.B. generated and analyzed the data and participated in drafting the manuscript. All authors had access to study data and approved the decision to submit the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Video 1.
Supplementary Video 2.
Supplementary Video 3.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sosnovski, K.E., Braun, T., Amir, A. et al. Reduced LHFPL3-AS2 lncRNA expression is linked to altered epithelial polarity and proliferation, and to ileal ulceration in Crohn disease. Sci Rep 13, 20513 (2023). https://doi.org/10.1038/s41598-023-47997-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-47997-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.