Abstract
Obesity induces a chronic inflammatory state associated with changes in adipose tissue macrophages (ATMs). Macrophage scavenger receptor 1 (MSR1) has been implicated in the regulation of adipose tissue inflammation and diabetes pathogenesis; however, reports have been mixed on the contribution of MSR1 in obesity and glucose intolerance. We observed increased MSR1 expression in VAT of obese diabetic individuals compared to non-diabetic and single nuclear RNA sequencing identified macrophage-specific expression of MSR1 in human adipose tissue. We examined male Msr1−/− (Msr1KO) and WT controls and observed protection from obesity and AT inflammation in non-littermate Msr1KO mice. We then evaluated obese littermate Msr1+/− (Msr1HET) and Msr1KO mice. Both Msr1KO mice and Msr1HET mice became obese and insulin resistant when compared to their normal chow diet counterparts, but there was no Msr1-dependent difference in body weight, glucose metabolism, or insulin resistance. Flow cytometry revealed no significant differences between genotypes in ATM subtypes or proliferation in male and female mice. We observed increased frequency of proliferating ATMs in obese female compared to male mice. Overall, we conclude that while MSR1 is a biomarker of diabetes status in human adipose tissue, in mice Msr1 is not required for obesity-associated insulin resistance or ATM accumulation.
Similar content being viewed by others
Introduction
Type 2 Diabetes (T2D) is a metabolic disorder characterized by insulin resistance and hyperglycemia and is a growing health concern. Obesity is the number one modifiable risk factor for T2D1,2. The association between obesity and T2D is well established, but the mechanisms that link the two remain incompletely understood. Adipose tissue is a key player in regulating insulin sensitivity and nutrient storage3. It is composed of adipocytes and non-adipocytes, including leukocytes such as adipose tissue macrophages (ATM)4. Under lean conditions, there is a balance between adipokines and anti-inflammatory cytokines to maintain metabolic homeostasis. Obesity induces adipose tissue dysfunction characterized by a state of chronic inflammation and adipocyte insulin resistance5,6,7. ATMs are a primary mediator of cytokine production in adipose tissue, producing pro-inflammatory cytokines that have been shown to inhibit glycogen synthesis8, decrease glucose uptake9 and impair insulin signaling10,11 impacting systemic insulin resistance. These findings along with animal models showing that impaired macrophage activation improved metabolism with obesity suggests that ATMs have a direct effect on obesity-induced insulin resistance.
However, many gaps remain in understanding the beneficial and detrimental effects of ATM functions and how these functions are regulated. Along with a quantitative increase in the number of ATMs with obesity and insulin resistance, the expression of macrophage scavenger receptor (MSR1/Scavenger receptor A) also increases in adipose tissue12 suggesting that MSR1 plays a role in promoting T2D. MSR1 expression in subcutaneous adipose tissue (SAT) is associated with insulin resistance in non-diabetic patients13. GWAS studies indicate associations between SNPs in MSR1 with waist-to-hip ratio and hip circumference adjusted for BMI14. The mechanism for this association is unknown.
MSR1 is a scavenger receptor prevalent in macrophages that binds many ligands, including low-density lipoproteins15. MSR1 is thought to aid in phagocytosis16,17 and promote inflammation in macrophages18,19. Msr1 has been shown to play a role in macrophage proliferation in peritoneal macrophages from Ldlr/− mice and in macrophages in atherosclerotic lesions in Apoe−/− mice20,21. Similarly, in adipose tissue, both M2-like resident ATMs (CD11c−) present in lean states and recruited M1-like proinflammatory ATMs (CD11c+) proliferate robustly with acute and chronic high-fat diet-induced obesity22,23. The persistent proliferation of CD11c+ ATMs contributes to the long-term effects of obesity on adipose tissue function even after weight loss24. The factors that regulate ATM proliferative signals are not known.
How or if MSR1 may play a role in ATM function is not well understood. Despite the clinical association between MSR1 expression and T2D, there is evidence from mouse studies suggesting that Msr1 may regulate beneficial macrophage functions to protect against obesity-induced insulin resistance. Msr1−/− ob/ob mice were found by Zhu et al. to be more insulin resistant and glucose intolerant compared to Msr1+/+ ob/ob mice25. Similar worse insulin resistance was observed in Msr1−/− mice fed HFD for 16 weeks. Cavallari et al.showed impaired glucose tolerance in obese (6-week HFD) Msr1−/− mice compared to Msr1+/+ mice26. On the other hand, Govaere et al.reported that obese Msr1−/− mice have increased weight gain, but are protected from obesity-induced insulin resistance, adipose tissue inflammation, and hepatic steatosis suggesting a pro-inflammatory role for Msr127.
Because obesity is associated with macrophage accumulation and chronic inflammation, Zhu et al. assessed ATMs to explain their metabolic phenotype in HFD Msr1+/+ and Msr1−/− mice and showed a decrease in resident CD11c− ATMs and an increase in proinflammatory CD11c+ ATMs in Msr1−/− ob/ob mice25. However, Callivari et al.did not note any significant differences in adipose tissue inflammation in their model and observed that administration of MSR1 agonists worsened glucose tolerance. Conversely, Zhu et al. suggested protective effects of Msr1 activation25,26. Govaere et al. showed that Msr1 promotes lipid accumulation and hepatic macrophage activation, suggesting a role for Msr1 in promoting obesity-induced inflammation. Furthermore, inhibition of Msr1 reduced fibrosis and macrophage activation in mouse and human liver, identifying Msr1 as a potential target for treating obesity-associated inflammation and liver damage27. Together, these results suggest that MSR1 may play a role in obesity-associated insulin resistance and adipose tissue inflammation, but the precise mechanisms are unclear.
We noted that weakness of all of these studies was that littermate controls were not utilized for comparisons and female mice were not assessed. Using littermate controls has been shown to be a more rigorous approach for analysis of genetically modified mice as the use of non-littermate parallel colonies can have epigenetic and environmental backgrounds that can skew results28. In diabetes research, the lack of littermate controls has been suggested to be a significant confounding variable in interpreting metabolic phenotypes in mice29.
To address these gaps and evaluate the hypothesis that MSR1 promotes ATM proliferation, we set out to examine the role for Msr1 in diet induced obesity in mice and humans. We observed associations with MSR1 expression in visceral adipose tissue (VAT) in obese diabetic patients and confirmed that MSR1 is highly restricted to ATMs in human adipose tissue in single nuclear RNAseq analysis. Initial studies suggested protection from diet induced obesity in Msr1−/− mice compared to non-littermate controls. However, comparison of littermate male and female mice did not demonstrate significant differences in obesity, glucose intolerance, or adipose tissue inflammation between genotypes. Our studies suggest that MSR1 is a strong ATM biomarker in human adipose tissue associated with T2D status, but our data does not support a functional role for Msr1 in regulating metabolic inflammation in mice.
Methods & materials
Animals and diet
Msr1+/− (Msr1HET) mice were mated with Msr1−/− (Msr1KO) mice (Jackson Laboratory; Strain 006,096) to produce experimental Msr1−/− pups and Msr1+/− littermate controls. Pups were weaned at 21 days and assessed for sex. Mice were separated into four experimental groups: 1) Msr1+/− on normal chow diet (ND) (10% fat; Research Diets: D12450J), 2) Msr1−/− mice on ND, 3) Ms1r+/− on a high-fat diet (HFD) (60% fat; Research Diets: D12492), 4) Msr1−/− on HFD. Each group was fed a ND until 8 weeks of age at which time groups 3 and 4 were switched to HFD. All groups had free access to food and water. Weight was obtained at the final endpoint. For each time point, each group of mice was euthanized, and adipose tissue was collected for analysis. For non-littermate control experiments, Msr1−/− (Msr1KO) mice (Jackson Laboratory; Strain 006,096) were compared with age-matched C57BL/6 J male mice. For non-littermate experiments, Msr1KO and WT mice were housed in separate cages. For littermate experiments, Msr1HET and Msr1KO littermate mice were housed in the same cages. All animal protocols were approved by the University of Michigan Institutional Animal Care and Use Committee. All methods were performed in compliance with ARRIVE guidelines, the NIH Public Health Service Policy on Human Care and Use of Laboratory Animals, and the NIH Guide for the Care and Use for Animals.
Glucose & insulin tolerance testing
Mice were fasted for 6 h and injected intraperitoneally (i.p.) with 1.0 g/kg of 10% glucose for the glucose tolerance test (GTT) or 1.0U/kg insulin for insulin tolerance test (ITT). Glucose levels were assessed at baseline and then every 15 min for 2 h using the Freestyle Freedom Lite glucometer and test strips.
Flow cytometry
Mice were euthanized and adipose tissue depots and liver were dissected at the end of the experiment. Adipose tissue was subjected to collagenase digestion to isolate the stromal vascular fraction (SVF) as described30. SVF was incubated with antibodies to identify macrophage subsets and proliferation and analyzed by flow cytometry on the BD Biosciences LSRFortessa™ cell analyzer as described30. The following conjugated antibodies were used: CD301 APC (Clone ER-MP23, BioRad, Cat# MCA2392A647), CD45 Pacific Blue (Clone 30-F11, eBiosciences, Cat# 48,045,182), CD64 PE (Clone X54-6/7.1.1, BD Biosciences, Cat# 558,455), CD11c APC-Cy7 (Clone N418, eBiosciences, Cat# 47,011,482), and Ki67 PeCy7 (Clone SolA15, eBiosciences, Cat#25,569,882). Single-stained, isotype, and unstained SVF were used as controls.
Gene expression
Total RNA was obtained from frozen mouse white adipose tissue via mechanical homogenization using the TissueMiser homogenizer (Fisher Scientific) and isolated using Qiagen’s Rneasy® Mini Kit (Cat# 74,106; Qiagen, Hilden, Germany). RNA was treated with DNase I (Cat# 79,254; Qiagen, Hilden, Germany)) and cDNA was prepared using 9.8-20 ng total RNA and Applied Biosciences™ High-Capacity cDNA Reverse Transcription Kit (Cat# 4,368,814; Thermo Fisher Scientific, Waltham, MA, USA). Transcript expression was measured using SYBR Power Green PCR Master Mix (Cat# 4,367,659; Thermo Fisher Scientific, Waltham, MA, USA) in a Quantstudio™ 3 real-time PCR cycler (Applied Biosystems by Thermo Fisher Scientific, Waltham, MA, USA). Target genes were compared to the geometric mean of the housekeeping gene, Arbp using the ΔΔCT method. Gene expression was analyzed in Msr1KO (n = 5) and Msr1HET mice (n = 3). Supplementary Table 1 lists primers for qPCR.
RNA sequencing analysis
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Analysis of MSR1 expression was performed using negative binomial regression model including sex, age and BMI in the model in R. The data used for the GTEX described in this manuscript are available at dbGaP accession number phs000424.vN.pN on 11/12/2022.
Results
MSR1 is macrophage-specific and is increased in visceral adipose tissue from obese diabetic humans
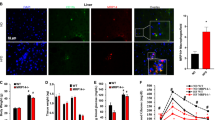
To assess the expression of MSR1 in human adipose tissue, we first interrogated the Genotype-Tissue Expression (GTEx) Project database for MSR1 expression in bulk RNA sequencing data. Adjusting for sex and age, MSR1 expression in both omentum/visceral adipose tissue (VAT) and subcutaneous adipose tissue (SAT) were significantly associated with BMI (VAT: Coefficient = 0.0280, p-value = 0.015; SAT Coefficient = 0.0370, p-value < 0.001) (Fig. 1a). We have previously published analysis of bulk RNA sequencing data from VAT and SAT from obese patient with and without diabetes mellitus (DM). Comparing non-DM (NDM) with DM patients, MSR1 expression was significantly increased in adipose tissue of DM patients. (Fig. 1b) Further stratifying the dataset by depot identified significantly higher MSR1 expression in SAT compared to VAT (Fig. 1c). Stratifying by depot and DM status, we observed higher expression of MSR1 in DM patients in VAT, but not SAT suggesting that MSR1 is a VAT specific biomarker of DM in humans (Fig. 1d).
MSR1 gene expression is increased in diabetic visceral adipose tissue in humans. (a) Correlation between MSR1 expression and BMI in omental/visceral (VAT; n = 354) and subcutaneous (SAT; n = 439) adipose tissue in GTEX. Negative binomial model used incorporating BMI, sex, and age in the model. (b–d) MSR1 expression from RNAseq data from independent cohort of VAT and SAT samples (n = 40). Corrected counts reported comparing (b) non-diabetic (NDM) and diabetic (DM), (c) VAT and SAT, and (d) depots stratified by DM status. *p-value < 0.05. ** p-value < 0.01. ***p-value < 0.001. (e) UMAP plots from single nuclear RNAseq data from human VAT and SAT identifying major cell types. MSR1 expression shown restricted to macrophages (MAC) cells. (f) Subcluster analysis of macrophages and T cells from single nuclear RNAseq analysis identifying lipid associated macrophages (LAM), TIM4 macrophages, and two types of MRC1 macrophages. MSR1 expression identified in all ATM subtypes.
To identify the cell types that express MSR1 in human adipose tissue, we interrogated a single nuclear RNAseq dataset we generated from obese human adipose tissue31. MSR1 expression was exclusive to adipose tissue macrophages (MAC) and not seen in other stromal cell types (Fig. 1e). Similar results were observed in interrogation of murine and human single nuclear RNA sequencing studies by Emont et al.32. Subset analysis of ATMs identified multiple subtypes including lipid-associated macrophages (LAM). MSR1 was expressed in all ATM subtypes and was not significantly enriched in one ATM type compared to others (Fig. 1f). This data indicates that MSR1 is a biomarker of all ATMs in human adipose tissue.
Msr1KO mice are protected from HFD-induced obesity compared to non-littermate controls
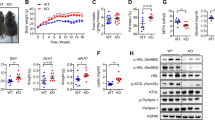
We performed experiments to compare the impact of high fat diet induced obesity on age matched WT and Msr1KO male mice maintained as parallel lines. After 12 weeks of HFD, Msr1KO mice had significantly less body weight and smaller fat pads compared to WT controls (Fig. 2a, b). While fasting glucose levels were similar, insulin tolerance tests demonstrated no significant differences in insulin sensitivity in Msr1KO male mice compared to controls (Fig. 2c). To assess whether Msr1 was required for obesity-induced adipose tissue inflammation, flow cytometry was performed on the SVF from three fat pads: epididymal white adipose tissue (eWAT), inguinal white adipose tissue (iWAT), and omental white adipose tissue (oWAT). Msr1KO mice had a lower frequency of CD45+ leukocytes in eWAT and iWAT, but more leukocytes were seen in Msr1KO oWAT (Fig. 2d). Msr1KO mice also had fewer total CD64+ ATMs in eWAT and oWAT, but not iWAT (Fig. 2e). When we assessed adipose tissue dendritic cells (ATDCs) and ATM subtypes we found that Msr1KO mice had an increase in CD64−CD11c+ ATDCs and resident CD64+CD11c− ATMs in eWAT compared to WT controls while inflammatory CD64+CD11c+ ATMs decreased (Fig. 2f, h). The frequency of resident CD64+CD11c− ATMs was also lower in oWAT, indicative of depot-specific differences. We observed fewer CD45+CD64+Ki67+ proliferating ATMs in eWAT and oWAT, but not iWAT from Msr1KO mice after 12 weeks of HFD (Fig. 2i). These results suggest that Msr1 is necessary for weight gain with HFD induced obesity and may be required to sustain obesity-associated inflammation. Our data also suggest a role for Msr1 in regulating adipose tissue inflammation via promoting ATM proliferation.
Male Msr1KO mice have decreased fat mass and inflammation compared to non-littermate wild-type controls. (a) Whole body weight for 12-week HFD fed Msr1KO (red) and C57BL6 wild type non-littermate controls (white). (b) Organ weight for Msr1KO (red) and WT non-littermate controls (black). (c) Insulin tolerance test (ITT) after 12 weeks of HFD. Flow cytometry analysis of (d) CD45+ leukocytes, (e) CD64+ ATMs, (f) CD64−CD11c+ ATDCs, (g) CD11c+ ATMs, (h) CD11c+ ATMs, and (i) CD64+Ki67+ ATMs in SVF after 12 weeks of HFD. n = 5/group. *p < 0.05. Statistical Analysis = T-test. Identical results observed when data was expressed as percent total SVF cells.
Msr1KO mice are not protected from HFD-induced obesity compared to Msr1HET littermate controls
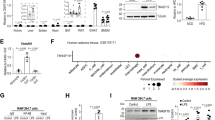
As these results contrasted with prior reports of worse insulin resistance in Msr1KO mice, to improve the rigor of the study, we compared Msr1KO mice and littermate Msr1HET controls. COVID-19 pandemic restrictions did not allow us to examine littermate Msr1HET, Msr1KO, and Msr1 + / + littermates and the lines were subsequently lost. To justify the use of Msr1HET mice as a control, qRT-PCR was performed from eWAT samples demonstrating loss of Msr1 expression in the KO mice and ~ 2 lower expression in HET mice compared to WT controls (Fig. 3a). Adipose tissue was harvested from experimental Msr1KO and Msr1HET littermate control male mice and analyzed for Msr1 expression, along with scavenger receptors Olr1 and Cd36, and cytokines Tnfα, IL-10, and IL1β by qPCR. Msr1KO mice had significantly less expression of Msr1 in eWAT than Msr1HET mice (Fig. 3a). Gene expression of Olr1, Cd36, Tnfα, IL-10, and IL1β was not different between the two groups. This data demonstrated that Msr1KO mice and Msr1HET mice are differentiated by Msr1 expression, making Msr1HET an appropriate control to explore the requirement of Msr1 in obesity-associated inflammation and insulin resistance.
Msr1 is not required for obesity-induced changes in glucose metabolism (a) Gene expression of Msr1, scavenger receptors, and cytokines in iWAT and eWAT by qPCR from male Msr1HET (black) and Msr1KO (red) male mice. n = 5/group. *p < 0.05; Statistical Analysis = t-test. WT mice were non-littermate HFD fed controls. (b) Whole body weight for 14-week male ND and HFD-fed Msr1HET and Msr1KO mice at the time of (c) glucose tolerance test (GTT). (d) Whole body weight for 15-week male HFD-fed mice at the time of (e) Insulin tolerance test (ITT). n = 3–5 per group. (f) Whole body weight for 16-week female HFD-fed Msr1HET (black) and Msr1KO (red) female mice. (g) Glucose tolerance test (GTT) and (h) insulin tolerance test (ITT) after 16 and 17 weeks of HFD, respectively. HFD Msr1HET (black), HFD Msr1KO (red). n = 4/group. Statistical Analysis for weight = Ordinary one-way ANOVA. ****p < 0.0001 between Msr1KO ND and HFD groups; Statistical Analysis for GTT/ITT = 2-way ANOVA multiple comparisons. **p < 0.05 between ND and HFD groups of the same genotype.
To evaluate the role of Msr1 on obesity-associated impaired glucose metabolism and insulin resistance, 8-week-old male Msr1KO and Msr1HET mice were fed HFD to induce obesity, then assessed by glucose (GTT) and insulin tolerance tests (ITT) at 14 and 15 weeks HFD, respectively. Both HFD groups gained significantly more weight than ND mice, but there were no differences in body weight between genotypes for ND or HFD-fed mice. (Fig. 3b). GTT showed that HFD-fed mice had worse glucose tolerance compared to ND fed mice but there were no differences between genotypes (Fig. 3c). HFD groups of mice were assessed for body weight and ITT at 15 weeks of HFD. Again, there was no difference in body weight or ITT responses between HFD Msr1HET and Msr1KO mice (Fig. 3d, e).
We also assessed response to HFD in female Msr1HET and Msr1KO mice. Both genotypes had similar weights after 16 weeks of HFD (Fig. 3f). Similar to male mice, there were no significant differences in GTT or ITT between genotypes after HFD induced obesity (Fig. 3g, h). This data suggests that Msr1 is not required for weight gain and insulin resistance during diet-induced obesity in either male or female mice.
Msr1KO is not required for ATM accumulation with HFD
We examined if Msr1 was required for obesity-induced ATM infiltration and proliferation. After 16 weeks of HFD, the SVF was isolated from fat pads and flow cytometry performed to quantify ATM subtypes and assess ATM proliferation. Both genotypes had similar body weights and there were no significant differences between spleen, eWAT, and liver weights between genotypes (Fig. 4a, b). Msr1KO mice had a trend for smaller iWAT fat pads compared to Msr1HET (p-value = 0.078). Flow cytometry analysis of SVF revealed no differences in the quantity of CD45 cells between Msr1-deficient mice and littermate controls in either eWAT or iWAT (Fig. 4c, d). Further analysis of the CD45+ leukocytes in the SVF revealed that Msr1KO and Msr1HET mice had the same frequency of total CD64+ ATMs, CD64+CD11c− resident ATMs, CD64+CD11c+ inflammatory ATMs, and CD64−CD11c+ ATCDs (Fig. 4e–f). We quantified proliferating ATMs in both genotypes by Ki67 staining. We analyzed CD64+Ki67+, CD64+CD11c−Ki67+, and CD64+CD11c+Ki67+ ATMs from eWAT and iWAT and no significant differences in proliferating ATMs were observed between genotypes (Fig. 4g). We observed that there was a significant higher proportion of proliferating CD64+CD11c+Ki67+ ATMs in iWAT compared to eWAT in Msr1KO.
Msr1 is not required for ATM infiltration during obesity in obese male mice. (a) Whole body weight and (b) organ weight for 16-week male HFD-fed Msr1HET (black) and Msr1KO (red) mice. Flow cytometry analysis of CD45+ cells in the SVF (c, d); ATM subtypes (e–f); and proliferating ATMs (g) in eWAT and iWAT. (h) Representative flow cytometry graphs. n = 3–5/group. ***p = 0.0005. Statistical Analysis = 2-way ANOVA multiple comparisons.
Similar analyses in female mice revealed no difference in body weight or organ weight after 21 weeks of HFD. (Fig. 5a, b). Quantitation of ATMs by flow cytometry after 21 weeks of HFD demonstrated no differences in total CD45+ leukocytes in eWAT or iWAT between genotypes (Fig. 5c, d). HFD fed female mice fail to induce significant CD11c+ ATMs (Fig. 5e–f) compared to male mice, but there were no differences between genotypes observed in total or subset ATM content. Similar levels of Ki67+ proliferating ATMs were observed between genotypes (Fig. 5g). However, when compared with male mice, HFD female mice had a greater frequency of proliferating total CD64+Ki67+ ATMs and subtypes in both iWAT and eWAT (Fig. 5h). Overall, these observations indicate that Msr1 is not required for obesity-associated adipose tissue inflammation and ATM infiltration and demonstrates that ATM proliferation is more robust in females compared to males.
Msr1 is not required for ATM infiltration during obesity in obese female mice. (a) Whole body weight and (b) organ weight after 21 weeks of HFD, at the time of euthansia. Flow cytometry analysis showing the frequency of (c, d) CD45+ cells in SVF, (e–f) %CD45 cells, and (g) proliferating ATMs in eWAT and iWAT from female Msrt1HET and Msr1KO mice. (h) Quantitation of proliferating ATMs in HFD fed male and female Msr1HET mice. (i) Representative flow cytometry graphs for (e–f). n = 3–5 per group. Statistical Analysis = t-test.
Discussion
This study aimed to elucidate the role of Msr1 in insulin resistance and adipose tissue inflammation during obesity. The premise of the study is based on our initial observations that that MSR1 is increase in the VAT of obese, diabetic humans and correlates with BMI in GTEX in both VAT and SAT. Analysis of published single nuclear RNA sequencing datasets demonstrate that MSR1 expression is highly restricted to ATMs and may be a biomarker for total ATM content in human adipose tissue. We were able to observe increased ATM proliferation in HFD fed female mice compared to males by flow cytometry analysis. We observed this in both subcutaneous and visceral adipose tissue depots and related to increased proliferation in all subtypes of ATMs. This result support previous observations of increased female ATM proliferation using Ki67 staining in adipose tissue sections with lipolysis33. Why female ATM proliferation is more prominent in males is unclear. It is possible that an increased proliferative capacity in female mice is related to suppression in obesity-induced adipose tissue inflammation compared to males34.
Because we did not observe changes in ND male mice and because female mice do not exhibit the same degree of obesity as male mice33,35, experiments in female mice were only carried out in HFD-fed mice. Our observation of a lack of Cd11c + inflammatory ATMs in female adipose tissue despite significant hypertrophy is consistent with prior studies showing a blunted recruitment of ATMs in female mice. While initial cohorts suggested protection from obesity and impaired ATM proliferation in male Msr1KO mice this was not replicated in littermate controls. We were unable to replicate prior studies suggesting that Msr1-deficient mice were more susceptible to inflammation with HFD induced obesity and instead initially observed that Msr1-deficient mice were protected from HFD-induced obesity and adipose tissue inflammation. The difference in weight gain observed between Msr1KO and WT non-littermate controls may be the primary cause for the protection from obesity-induced inflammation observed in Msr1KO mice. Protection from DIO is a common feature in many mouse models where macrophage genes are knocked out including other scavenger receptors such as CD36 and may be due to increased energy expenditure36,37,38. The differences we observed in obesity development in our non-littermate experiments could be attributed to differences in the microbiome as a result of the separate housing conditions.
To ensure a more rigorous experimental design, we compared Msr1KO mice with Msr1HET littermate controls. While there is no standard model for HFD feeding to induce obesity, C57BL6 mice exhibit significant weight gain and insulin resistance between 16 and 22 weeks of HFD39. Since we observed differences in body weight between the genotypes in the non-littermate studies, we extended the duration of HFD feeding to 16 weeks to ensure weight gain in the littermate studies With this experimental design we observed that both male and female Msr1HET and Msr1KO mice exhibited the same degree of HFD-induced weight gain, insulin resistance, and adipose tissue inflammation. Our findings suggest that Msr1 does not play a significant role in controlling obesity-associated insulin resistance and inflammation.
One limitation of the studies may be the use of Msr1HET littermates as the control group as it is possible that Msr1HET mice with half the expression of WT mice. Because Msr1KO mice had significantly less Msr1 expression in their adipose tissue compared to their Msr1HET littermates, we still feel this rigorously tests the requirement for Msr1 in HFD induced obesity. However, it is possible that Msr1HET have a phenotype that differs from WT mice although no groups have reported abnormalities in Msr1HET mice. COVID-19 pandemic restrictions did not allow us to examine littermate Msr1HET, Msr1KO, and Msr1 + / + littermates. We also attempted to use commercially available antibodies to verify Msr1 expression but were unable to get these reagents to detect Msr1 by immunofluorescence or flow cytometry (data not shown).
Using littermate controls instead of mice from a parallel but distinct colony is recommended in metabolic studies to ensure the epigenetic and environmental backgrounds are comparable28. Having the appropriate control is necessary to determine the phenotype of genetically modified strains and can be a significant source of variation in metabolic studies. The Msr1KO mice were developed from 129SV embryonic stem cells injected into C57BL/6 J blastocysts and are backcrossed to C57BL/6JIco mice. Studies found that 129 X C57BL/6 mice display spontaneous autoimmunity, and in mutant mice backcrossed to this strain the target gene is likely influenced by surrounding 129SV genes40,41. Therefore, it is possible that the effects of the mixed background are significant and need to be controlled for more rigorously. However, we did conduct experiments to directly test if Msr1HET littermates had an altered phenotype compared to C57BL/6 WT mice however, the results we obtained in this study are all similar to our experience with HFD feeding in C57Bl/6 mice in our colony25,26.
Our inability to replicate findings with Msr1KO mice from other studies may be due to other factors that influence mouse metabolism. A recent large-scale study comparing genetically identical mice (C57BL/6 J) housed at four separate Mouse Metabolic Phenotyping Centers (MMPC) demonstrated substantial variation in the response to identical HFD feeding paradigms based on location42. Significant differences in site-specific metabolic rates were observed suggesting that location-specific differences in housing conditions, equipment, microbiome, or other factors may contribute to variation in responses to HFD induced obesity. Regression analysis identified body composition, activity, photoperiod, diet and acclimation conditions contributed the most to variance in energy expenditure. Single nuclear RNA sequencing from human adipose tissue demonstrates that MSR1 is highly specific to ATMs. The association between MSR1 expression and DM status may be proportional to ATM content which is increased in DM patients based on our recent results and other studies43. We attempted to validate MSR1 protein expression in mouse adipose tissue by immunofluorescence and flow cytometry but failed to obtain specific staining with commercially available MSR1 antibodies.
Overall, our experiments do not support a functional role for MSR1 in obesity-induced adipose tissue inflammation despite being a biomarker for ATM content. Our initial hypothesis of a role for MSR1 in adipose tissue inflammation was based on prior research supporting roles for MSR1 in the regulation of antigen presentation44 and in lipid uptake in macrophages in the setting of fatty liver disease27. Given differences between mice and humans in ATM function, future studies should evaluate potential functional roles for MSR1 activity in human ATM function.
Data availability
The single nuclear RNA sequencing datasets generated and/or analyzed during the current study are available in the Single Cell Portal at #SCP190331. The data used for the GTEX described in this manuscript are available at dbGaP accession number phs000424.vN.pN on 11/12/2022.
References
Grant, B., Sandelson, M., Agyemang-Prempeh, B. & Zalin, A. Managing obesity in people with type 2 diabetes. Clin. Med. (Lond) 21, e327–e231. https://doi.org/10.7861/clinmed.2021-0370 (2021).
Barnes, A. S. The epidemic of obesity and diabetes: trends and treatments. Tex Heart Inst. J. 38, 142–144 (2011).
Smith, U. & Kahn, B. B. Adipose tissue regulates insulin sensitivity: role of adipogenesis, de novo lipogenesis and novel lipids. J. Intern. Med. 280, 465–475. https://doi.org/10.1111/joim.12540 (2016).
Esteve Rafols, M. Adipose tissue: cell heterogeneity and functional diversity. Endocrinol Nutr. 61, 100–112. https://doi.org/10.1016/j.endonu.2013.03.011 (2014).
Russo, L. & Lumeng, C. N. Properties and functions of adipose tissue macrophages in obesity. Immunology 155, 407–417. https://doi.org/10.1111/imm.13002 (2018).
Lumeng, C. N. Innate immune activation in obesity. Mol. Aspects Med. 34, 12–29. https://doi.org/10.1016/j.mam.2012.10.002 (2013).
Lumeng, C. N. & Saltiel, A. R. Inflammatory links between obesity and metabolic disease. J. Clin. Invest. 121, 2111–2117. https://doi.org/10.1172/JCI57132 (2011).
Kanemaki, T. et al. Interleukin 1beta and interleukin 6, but not tumor necrosis factor alpha, inhibit insulin-stimulated glycogen synthesis in rat hepatocytes. Hepatology 27, 1296–1303. https://doi.org/10.1002/hep.510270515 (1998).
Carey, A. L. et al. Interleukin-6 increases insulin-stimulated glucose disposal in humans and glucose uptake and fatty acid oxidation in vitro via AMP-activated protein kinase. Diabetes 55, 2688–2697. https://doi.org/10.2337/db05-1404 (2006).
Uysal, K. T., Wiesbrock, S. M., Marino, M. W. & Hotamisligil, G. S. Protection from obesity-induced insulin resistance in mice lacking TNF-alpha function. Nature 389, 610–614. https://doi.org/10.1038/39335 (1997).
Jager, J., Gremeaux, T., Cormont, M., Le Marchand-Brustel, Y. & Tanti, J. F. Interleukin-1beta-induced insulin resistance in adipocytes through down-regulation of insulin receptor substrate-1 expression. Endocrinology 148, 241–251. https://doi.org/10.1210/en.2006-0692 (2007).
Santiago-Fernández, C. et al. in Society for Endocrinology BES Vol. 41 (Endocrine Abstracts, 2016).
Rasouli, N. et al. Association of scavenger receptors in adipose tissue with insulin resistance in nondiabetic humans. Arterioscler. Thromb. Vasc. Biol. 29, 1328–1335. https://doi.org/10.1161/ATVBAHA.109.186957 (2009).
Tachmazidou, I. et al. Whole-genome sequencing coupled to imputation discovers genetic signals for anthropometric traits. Am. J. Hum. Genet. 100, 865–884. https://doi.org/10.1016/j.ajhg.2017.04.014 (2017).
Brown, M. S., Basu, S. K., Falck, J. R., Ho, Y. K. & Goldstein, J. L. The scavenger cell pathway for lipoprotein degradation: specificity of the binding site that mediates the uptake of negatively-charged LDL by macrophages. J. Supramol. Struct. 13, 67–81. https://doi.org/10.1002/jss.400130107 (1980).
Kelley, J. L., Ozment, T. R., Li, C., Schweitzer, J. B. & Williams, D. L. Scavenger receptor-A (CD204): a two-edged sword in health and disease. Crit. Rev. Immunol. 34, 241–261. https://doi.org/10.1615/critrevimmunol.2014010267 (2014).
Zhang, Z. et al. Scavenger receptor A1 attenuates aortic dissection via promoting efferocytosis in macrophages. Biochem. Pharmacol. 168, 392–403. https://doi.org/10.1016/j.bcp.2019.07.027 (2019).
Guo, M. et al. Triggering MSR1 promotes JNK‐mediated inflammation in IL‐4‐activated macrophages. EMBO J. https://doi.org/10.15252/embj.2018100299 (2019).
Kong, F. Q. et al. Macrophage MSR1 promotes the formation of foamy macrophage and neuronal apoptosis after spinal cord injury. J. Neuroinflammat. 17, 62. https://doi.org/10.1186/s12974-020-01735-2 (2020).
Robbins, C. S. et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med. 19, 1166–1172. https://doi.org/10.1038/nm.3258 (2013).
Sakai, M. et al. The scavenger receptor serves as a route for internalization of lysophosphatidylcholine in oxidized low density lipoprotein-induced macrophage proliferation. J. Biol. Chem. 271, 27346–27352. https://doi.org/10.1074/jbc.271.44.27346 (1996).
Muir, L. A. et al. Frontline science: rapid adipose tissue expansion triggers unique proliferation and lipid accumulation profiles in adipose tissue macrophages. J. Leukoc. Biol. 103, 615–628. https://doi.org/10.1002/JLB.3HI1017-422R (2018).
Amano, S. U. et al. Local proliferation of macrophages contributes to obesity-associated adipose tissue inflammation. Cell Metab. 19, 162–171. https://doi.org/10.1016/j.cmet.2013.11.017 (2014).
Zamarron, B. F. et al. Macrophage proliferation sustains adipose tissue inflammation in formerly obese mice. Diabetes 66, 392–406. https://doi.org/10.2337/db16-0500 (2017).
Zhu, X. et al. Deletion of class A scavenger receptor deteriorates obesity-induced insulin resistance in adipose tissue. Diabetes 63, 562–577. https://doi.org/10.2337/db13-0815 (2014).
Cavallari, J. F. et al. Targeting macrophage scavenger receptor 1 promotes insulin resistance in obese male mice. Physiol. Rep. 6, e13930. https://doi.org/10.14814/phy2.13930 (2018).
Govaere, O. et al. Macrophage scavenger receptor 1 mediates lipid-induced inflammation in non-alcoholic fatty liver disease. J. Hepatol. 76, 1001–1012. https://doi.org/10.1016/j.jhep.2021.12.012 (2022).
Holmdahl, R. & Malissen, B. The need for littermate controls. Eur. J. Immunol. 42, 45–47. https://doi.org/10.1002/eji.201142048 (2012).
Alquier, T. & Poitout, V. Considerations and guidelines for mouse metabolic phenotyping in diabetes research. Diabetologia 61, 526–538. https://doi.org/10.1007/s00125-017-4495-9 (2018).
Cho, K. W., Morris, D. L. & Lumeng, C. N. Flow cytometry analyses of adipose tissue macrophages. Methods Enzymol. 537, 297–314. https://doi.org/10.1016/B978-0-12-411619-1.00016-1 (2014).
Strieder-Barboza, C. et al. Single-nuclei transcriptome of human AT reveals metabolically distinct depot-specific adipose progenitor subpopulations. bioRxiv 102, 1032. https://doi.org/10.1101/2022.06.29.496888 (2022).
Emont, M. P. et al. A single-cell atlas of human and mouse white adipose tissue. Nature 603, 926–933. https://doi.org/10.1038/s41586-022-04518-2 (2022).
Varghese, M. et al. Sex Differences in inflammatory responses to adipose tissue lipolysis in diet-induced obesity. Endocrinology 160, 293–312. https://doi.org/10.1210/en.2018-00797 (2019).
Varghese, M. et al. Monocyte trafficking and polarization contribute to sex differences in meta-inflammation. Front. Endocrinol. 13, 826320. https://doi.org/10.3389/fendo.2022.826320 (2022).
Arcones, A. C., Cruces-Sande, M., Ramos, P., Mayor, F. Jr. & Murga, C. Sex differences in high fat diet-induced metabolic alterations correlate with changes in the modulation of GRK2 levels. Cells 8, 1464 (2019).
Ma, J. et al. CD226 knockout alleviates high-fat diet induced obesity by suppressing proinflammatory macrophage phenotype. J. Transl. Med. 19, 477. https://doi.org/10.1186/s12967-021-03150-4 (2021).
Cai, L., Wang, Z., Ji, A., Meyer, J. M. & van der Westhuyzen, D. R. Scavenger receptor CD36 expression contributes to adipose tissue inflammation and cell death in diet-induced obesity. PLoS One 7, e36785. https://doi.org/10.1371/journal.pone.0036785 (2012).
Desai, H. R. et al. Macrophage JAK2 deficiency protects against high-fat diet-induced inflammation. Sci. Rep. 7, 7653. https://doi.org/10.1038/s41598-017-07923-0 (2017).
Surwit, R. S., Kuhn, C. M., Cochrane, C., McCubbin, J. A. & Feinglos, M. N. Diet-induced type II diabetes in C57BL/6J mice. Diabetes 37, 1163–1167. https://doi.org/10.2337/diab.37.9.1163 (1988).
Bygrave, A. E. et al. Spontaneous autoimmunity in 129 and C57BL/6 mice-implications for autoimmunity described in gene-targeted mice. PLoS Biol. 2, E243. https://doi.org/10.1371/journal.pbio.0020243 (2004).
Kanagawa, O., Xu, G., Tevaarwerk, A. & Vaupel, B. A. Protection of nonobese diabetic mice from diabetes by gene(s) closely linked to IFN-gamma receptor loci. J. Immunol. 164, 3919–3923. https://doi.org/10.4049/jimmunol.164.7.3919 (2000).
Corrigan, J. K. et al. A big-data approach to understanding metabolic rate and response to obesity in laboratory mice. Elife https://doi.org/10.7554/eLife.53560 (2020).
Muir, L. A. et al. Human CD206+ macrophages associate with diabetes and adipose tissue lymphoid clusters. JCI Insight 7, e146563. https://doi.org/10.1172/jci.insight.146563 (2022).
Yi, H. et al. Suppression of antigen-specific CD4+ T cell activation by SRA/CD204 through reducing the immunostimulatory capability of antigen-presenting cell. J. Mol. Med. (Berl) 90, 413–426. https://doi.org/10.1007/s00109-011-0828-1 (2012).
Funding
The authors are funded by grants from the National Institutes of Health DK122724 (S.N.) DK129636 (L.M.) and DK090262 (C.L.).
Author information
Authors and Affiliations
Contributions
S.N., J.D., and L.M. designed, performed, analyzed, and interpreted the data. S.N., L.M. and C.L. designed, prepared data for presentation, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nance, S.A., Muir, L., Delproprosto, J. et al. MSR1 is not required for obesity-associated inflammation and insulin resistance in mice. Sci Rep 13, 2651 (2023). https://doi.org/10.1038/s41598-023-29736-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29736-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.