Abstract
Abnormal proline-rich protein 11 (PRR11) expression is associated with various tumors. However, there are few reports concerning PRR11 with prognostic risk, immune infiltration, or immunotherapy of bladder urothelial carcinoma (BLCA). This study is based on online databases, such as Oncomine, GEPIA, HPA, LinkedOmics, TIMER, ESTIMATE and TISIDB, and BLCA data downloaded from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus, we employed an array of bioinformatics methods to explore the potential oncogenic roles of PRR11, including analyzing the relationship between PRR11 and prognosis, tumor mutational burden (TMB), microsatellite instability, and immune cell infiltration in BLCA. The results depict that PRR11 is highly expressed in BLCA, and BLCA patients with higher PRR11 expression have worse outcomes. In addition, there was a significant correlation between PRR11 expression and TMB and tumor immune infiltration. These findings suggest that PRR11 can be used as a potential marker for BLCA patient assessment and risk stratification to improve clinical prognosis, and its potential regulatory mechanism in the BLCA tumor microenvironment and targeted therapy is worthy of further investigation.
Similar content being viewed by others
Introduction
Bladder cancer mainly includes urothelial (transitional cell carcinoma), squamous cell carcinoma, and adenocarcinoma. Among them, urothelial carcinoma is the most common, accounting for more than 90% of the total prevalence of bladder cancer1. It has been identified that cigarette smoke2, intake of aristolochic acid3, arsenic exposure4,5, and occupational exposure to aromatic amines6,7 have significant effects on tumorigenesis of BLCA. BLCA is the top ten cancer type and the second most prevalent genitourinary system tumor worldwide, with approximately 429,000 new cases and 165,000 deaths annually8. Despite advances in surgical techniques and postoperative management, the clinical outcomes of BLCA patients have not improved much over the past few decades due to the lack of sensitive markers9. Before diagnosing and treating BLCA, it is necessary to determine whether the cancer is muscle-invasive or non-muscle invasive. According to the clinical characteristics of the two, muscle-invasive cancer requires radical cystectomy, while non-muscle invasive cancer can be excised locally using minimally invasive surgery, followed by routine intravesical chemotherapy. The prognosis of 70% of cases with non-muscular invasive BLCA improves after treatment with transurethral resection (TURB) and intravesical chemotherapy, immunotherapy with Bacillus Calmette-Guérin10. However, 40% of treated patients will develop muscle-invasive BLCA within five years11. Patients with muscular invasive BLCA have a higher risk of long-distance metastasis and a worse prognosis than those with non-muscular invasive BLCA12. Thus, discovering specific early detection markers and therapeutic targets is the key to improving the survival rate of BLCA patients.
Immune cell infiltration is an important component of the tumor microenvironment (TME), and it plays a significant role in the initiation, progression, and clinical treatment of tumors13,14,15. Changes in the immune microenvironment may affect clinical outcomes in various tumors, such as melanoma, lung cancer, breast cancer, and muscle-invasive bladder cancer16. It has been reported that immune cells and related genes in the BLCA microenvironment promote tumor progression19, and the infiltrating characteristics of macrophages in BLCA may affect T-cell tolerance, thus altering patient outcomes17,18. Considering the current development of various cellular immunotherapy and targeted therapies as well as their clinical application trends, it is instructive to clarify the characteristics of immune infiltration and internal regulation mechanisms of various types of neoplastic disease.
The PRR11 gene on chromosome 17q22 encodes a proline-rich protein with a zinc finger domain and a bivalent nuclear localization signal20. Since its discovery in 2013, the relationship between this gene and tumors has been getting closer, and reports have revealed that it is related to the poor outcomes of various tumor types21, such as ovarian carcinoma22, breast cancer23, non-small cell lung cancer24, colorectal cancer25, and pancreatic cancer26. In addition, data indicate that it is involved in the epithelial-mesenchymal transition (EMT) process27. Therefore, we hypothesize that PRR11 may have diagnostic, prognostic, and targeted therapy applications in various tumor types. When performing an in-depth analysis of the full-gene difference of BLCA, our team discovered that PRR11 is a gene with a notable difference. Lin et al. also suggested that PRR11 may increase risk scores in bladder cancer patients28. Because there are few reports on the relationship between PRR11 and BLCA, we have conducted further research on this in the follow-up.
In this study, we analyzed and verified the expression of PRR11 in BLCA using GEPIA, Oncomine, TCGA, and other databases before calculating its significance in the clinical prognosis of patients using various statistical methods. Using Timer, ESTIMATE, TISIDB, and LinkedOmics database, we also clarified the primary biological function of PRR11 and its potential impact on BLCA immune infiltration. In conclusion, our findings suggested a significant prognostic value of PRR11 and a potentially promising target for immunotherapeutic strategies in BLCA.
Materials and methods
Data acquisition and processing
We used the TCGA database (https://genome-cancer.ucsc.edu/) to collect RNA-seq data and clinical information from 411 BLCA projects, including 19 cases of matching adjacent tissues. The downloaded data format is level 3 HTSeq-fragments per kilobase per million (FPKM). We also downloaded TPM (transcripts per million) format RNA-seq data of 9 normal human bladder tissues in Genotype-Tissue Expression (GTEx) database that uniformly processed by Toil process from UCSC Xena (https://xenabrowser.net/datapages/)29. The gene expression profiling data sets, GSE48276(n = 116) and GSE13507(n = 256), were obtained from GEO database (https://www.ncbi.nlm.nih.gov/gds).
We used R (v4.1.0) to generate the receiver operating characteristic (ROC) curve and analyze the expression of PRR11 in 19 paired samples in TCGA-BLCA dataset, and the pROC and “ggplot2” R package were used for visualization. The RNAseq data in the FPKM format of the paired samples were compared after the log2 conversion. The “survival” package was used for statistical analysis of survival data, the “survminer” package for visualization. The expression of PRR11 protein in bladder tissue was analyzed based on the immunohistochemistry data from HPA (Human Protein Atlas) database (https://www.proteinatlas.org/).
Genome-wide analysis of differential gene
Limma package (version: 3.40.2) of R software was used to study the differential expression of mRNAs in BLCA . The adjusted P-value was analyzed to correct for false positive results in TCGA or GTEx. “Adjusted P < 0.05 and Log (Fold Change) > 1 or Log (Fold Change) < − 1” were defined as the thresholds for the screening of differential expression of mRNAs (Supplementary Fig. 1). We selected genes with Log (Fold Change) > 2.5, P and adjusted P < 0.001 from the screening results, then set PRR11 as the research target.
Oncomine database analysis
We used Oncomine (https://www.oncomine.org/resource/login.html) to identify the expression level of PRR11 in various tumors30. The threshold was set at a P value of 1e-5, a multiple change of 1.5 and the gene rank of top 5%.
GEPIA database analysis
Gene expression profiling interactive analysis (GEPIA) is an interactive web to analyze the RNA sequencing expression, including 9736 tumors and 8587 normal samples from TCGA and the GTEx projects (http://gepia.cancer-pku.cn/index.html)31. Here, we used GEPIA to re-analyze and verify the expression of PRR11, and produced a total survival (OS) map of PRR11 related genes based on log-rank test in BLCA. In addition, we explored the correlation between PRR11 expression and gene markers of tumor infiltrating immune cells through GEPIA database32, and produced the scatter plots of PRR11 expression between a pair of user defined genes in BLCA.
Correlation of PRR11 expression with tumor mutation burden, tumor microsatellite instability, and mismatch repair gene expression
TMB and MSI scores were determined for all samples based on mRNA-seq data downloaded from TCGA (https://tcga.xenahubs.net), and Spearman’s correlation analysis was used to describe the correlation. Results are presented as scatter plot, produced by the R-package “ggstatspolt”. Expression profile data from TCGA were used to evaluate the levels of the mismatch repair (MMR) genes, including MutL homologous gene (MLH1), MutS homologous gene 2 (MSH2), MSH6, postmeiotic segregation increased 2 (PMS2), epithelial cell adhesionmolecule (EPCAM), in BLCA and determined the correlation between levels of MMR gene expression and PRR11. Data were visualized as a heatmap by the R-packages "ggstataplot".
Relationship between PRR11 and immunity
The correlation between PRR11 expression and immune cell infiltration in BLCA was explored by a gene module in the Tumor Immune Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/)33. The TIMER database contains 32 cancers from the TCGA. The immune cells in the TIMER database include CD4+ T cells, CD8+ T cells, B cells, neutrophils, macrophages, and dendritic cells.
The TISIDB database (http://cis.hku.hk/TISIDB) is a web portal for tumour and immune system interaction, which integrates multiple heterogeneous data types. They pertain to 988 reported immune-relatedanti-tumour genes, high-throughput screening techniques, molecular profiling and para-cancerous multi-omics data, as well as numerous resources for immunological data gathered from seven public databases34. In this study, TISIDB provided us associations for PRR11 with Th1 and Th2 lymphocytes.
Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) database (https://bioinformatics.mdanderson.org/public-software/estimate/) is an algorithm using gene expression data to infer tumor purity and the degree of infiltration of immune cells into tumors35. The immunescore and tumorpurity score of each patient in datasets was calculated by the R package “estimate”, and visualized by "ggplot2" R package.
Linked omics database analysis
LinkedOmics is publicly available portal that includes multi-omics data from all 32 TCGA Cancer types (http://www.linkedomics.org/login.php)36. The differentially expressed genes related to PRR11 were screened from the TCGA BLCA cohort through the LinkFinder module in the database, and the correlation of results was tested by the Spearman test and presented respectively in scatter plot and heat maps. Function module analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was performed by the gene set enrichment analysis (GSEA) in the LinkInterpreter module37,38,39.
Statistical analysis
Wilcoxon rank sum test, Chi-square test, Fisher exact test and logistic regression were used to analyze the relationship between clinicopathological characteristics and PRR11 expression. The Kaplan–Meier method was used to calculate the OS and disease-free survival (DFS). Cox proportional hazards model was used for univariate and multivariate analysis to estimate the association between PRR11 expression and clinical characteristics, OS. P value less than 0.05 is considered statistically significant. SPSS15.0 was used for statistical analysis.
Ethical approval
TCGA and GEO belong to public databases. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for research and publish relevant articles. Our study is based on open source data, so there are no ethical issues and other conflicts of interest.
Results
Increased PRR11 expression in BLCA
We initially used the Oncomine database to analyze the expression differences of PRR11 mRNA between cancer and normal tissues. The analysis demonstrated that the expression level of PRR11 mRNA was significantly increased in BLCA (Fig. 1a), which was also verified in the GEPIA database (Fig. 1b). PRR11 is also up-regulated in other types of cancer, such as breast, head and neck, and lung cancer, and its expression in sarcoma is particularly striking, which may be a new discovery. We then used 19 pairs of BLCA tissues and matched non-cancer tissues from the TCGA database to demonstrate that the comparison results were still accurate (Fig. 1c). Through the HPA database, we confirmed that the expression level of PRR11 protein in BLCA is still greater than that of normal tissues (Fig. 1d). In addition, we used the ROC curve to analyze the potential effectiveness of distinguishing BLCA from non-tumor tissues by PRR11 expression (Fig. 1e). The results demonstrated that the area under the curve (AUC) of PRR11 was 0.877, indicating that PRR11 can be used as an ideal biomarker to differentiating BLCA from non-tumorous tissues.
PRR11 expression is increased in BLCA. (a) Increased or decreased PRR11 in datasets of different cancers compared with normal tissues in the Oncomine database. Red represents high expression, blue represents low expression, and the shade of color depends on the degree of expression. (b) PRR11 expression in BLCA from TCGA database were determined by GEPIA. (c) The expression of PRR11 in 19 pairs of BLCA tissues and matched non-cancer tissues from the TCGA database. (d) The expression of PRR11 in normal and BLCA tissue sections from the HPA database. (e) ROC curve showed the efficiency of PRR11 expression level to distinguishing BLCA from non-tumor tissue in TCGA database. X-axis represents false positive rate, and Y-axis represents true positive rate. *P < 0.05, ***P < 0.001.
Prognostic potential of PRR11 in BLCA
We selected the optimal cut-off value of PRR11 (1.6025) based on the results of the ROC curve and divided 402 BLCA samples from the TCGA database into high expression and low expression groups. Kaplan–Meier analysis was used to explore the correlation between PRR11 expression and OS and DFS of BLCA patients. The results revealed that the poor prognosis in BLCA was significantly correlated with the high expression of PRR11 (Fig. 2a,b). In addition, the two datasets GSE48276 and GSE13507 were used to further validate the prognostic value of PRR11 (Fig. 2c–e). These results indicated that PRR11 has considerable prognostic value for BLCA.
Kaplan‐Meier survival curves were used to analyze the correlation between gene alterations in PRR11 and OS, as well as the RFS of BLCA patients. In TCGA-BLCA dataset, high PRR11 expression was correlated with poor OS (a) and RFS (b). Patients with higher PRR11 expression level had higher OS time in the GSE13507 (c) and GSE48276 dataset (e). (d) ROC curve showed the efficiency of PRR11 expression level to distinguishing BLCA from non-tumor tissue in GSE48276 dataset. X-axis represents false positive rate, and Y-axis represents true positive rate.
Correlation of PRR11 expression with clinicopathological factors in BLCA
The characteristics of samples were displayed in Table 1, in which 411 cases of BLCA with both clinical and gene expression data were collected from the TCGA database. The association between the level of PRR11 expression and the clinicopathological characteristics of BLCA patients was evaluated using the Chi-square test or Fisher's exact test, or Rank sum test. The results depicted that the expression level of PRR11 was significantly correlated with the patient's age, race, and pathological grade. In addition, the Logistic regression method demonstrated that PRR11 expression level is significantly correlated with pathological grade ( Table 2).
Cox univariate and multivariate analysis of prognostic factors in BLCA
Figure 3a displays the results of Cox univariate and multivariate analysis of OS in BLCA. The variables with P < 0.05 were PRR11, age T stage, N stage and M stage in the Cox univariate regression model. Multivariate analysis further demonstrated that PRR11 and N stage were independent prognostic factors in OS of BLCA patients (Fig. 3b).
Correlations of PRR11 expression with tumor mutation burden, tumor microsatellite instability and mismatch repair genes
Subsequently, considering the sensitivity of clinical immunotherapy, we investigated the correlation between the expression level of PRR11 and TMB, MSI, and MMR-related genes (MLH1, MSH2, MSH6, PMS2, and EPCAM). The results indicated that the expression of PRR11 in BLCA was positively correlated with TMB (Fig. 4a) but not significantly correlated with MSI (Fig. 4b). MMR gene expression is obviously and significantly positively correlated with PRR11 levels (Fig. 4c).
Correlation analysis of PRR11 expression and TMB (a)/MSI (b). The horizontal axis in the figure represents the expression distribution of the gene, and the ordinate is the expression distribution of the TMB/MSI score. The density curve on the right represents the distribution trend of the TMB/MSI score; the upper density curve represents the distribution trend of the gene; the value on the top side represents the correlation p value, correlation coefficient and correlation calculation method. (c) Heatmap illustrating the association between PRR11 expression and MMR genes. The gene correlation heatmap is realized by the R (v4.1.0) software package “ggstatsplot”. ***P < 0.001.
PRR11 coexpression network in BLCA
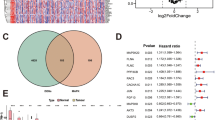
To further clarify the biological role of PRR11 in BLCA, we used the LinkedOmics database to explore the coexpression characteristics of PRR11 based on the BLCA samples from TCGA. The scatter plot illustrates that PRR11 is correlated with a total of 20,047 genes, of which 10,074 genes are positively correlated with it, and the remaining 9,973 genes are negatively correlated with it (Fig. 5a). We selected the top 50 genes that were positively and negatively associated with PRR11, and demonstrated their expression pattern as a heatmap (Fig. 5b,c). Further analysis of these 100 genes revealed that 37 of 50 positive correlation genes had unfavorable HR, and one was statistically significant (ANLN). In contrast, 34/50 genes negatively related to PRR11 had the possibility of being potential protective markers in BLCA patients, and 5 of them were statistically significant (IDUA, BATF, TNFRSF14, RILP, TMEM219). It was worth mentioning that SYNC, as a gene negatively correlated with PRR11, had unfavorable HR (Fig. 5d). The results of KEGG pathway enrichment analysis demonstrated that PRR11 co-expressed genes were mainly involved in the cell cycle, homologous recombination, microRNAs in cancer, RNA transport, DNA replication, and ErbB signaling pathway (Fig. 5e). GO term annotation (biological process, cellular component and molecular function) demonstrated that co-expressed genes of PRR11 are primarily associated with chromosome segregation, DNA replication, cell cycle, double-strand break repair, spindle organization and a series of biological processes surrounding nucleic acid, and also related to enzyme activities, such as Helicase and ATPase. (Supplementary Fig. 2). These findings imply that the PRR11 co-expression network may play an important role in the prognosis, initiation and progression of BLCA, but further follow-up studies are needed.
PRR11 coexpression network in BLCA from LinkedOmics database. (a) All genes associated with PRR11 in BLCA cohort, red dots positively correlated with PRR11, and green dots negatively correlated. Heat map of the top 50 genes positively (b) and negatively (c) correlated with PRR11 in BLCA. Red represents positively linked genes and blue represents negatively linked genes. The gene expression heatmap is realized by LinkedOmics database (http://www.linkedomics.org/login.php). (d) Survival map of the top 50 genes positively and negatively associated with PRR11 in BLCA. The survival heatmap is realized by GEPIA Database (http://gepia.cancer-pku.cn/index.html). (e) PRR11 of KEGG pathway enrichment analysis in BLCA cohort.
Association between PRR11 with immune infiltration
We used the TIMER database to investigate the relationship between PRR11 and the level of immune infiltration in tumors, given the significance of this parameter. The results revealed that the expression level of PRR11 and the infiltration level of CD8+ T cell, macrophage, neutrophil , and dendritic cells in BLCA were significantly positively correlated (Fig. 6). By analyzing the TISIDB database, we also found that the infiltration level of Th2 cell was significantly positively correlated with the PRR11 expression, whereas Th1 did not have any significant correlations with the expression of PRR11 (Fig. 7a,b).
Then, we used the ESTIMATE algorithm to determine whether the expression of PRR11 was associated with the infiltration level of total immunity in BLCA. The calculation results demonstrated that in the two datasets of TCGA-BLCA and GSE48276, PRR11 and Immunescore had a significant negative correlation, and a significant positive correlation with tumor purity (Fig. 8a–d). The prognostic analysis also revealed that patients with high immune scores had better overall survival time than patients with low scores, whereas patients with high tumor purity scores had a worse prognosis (Fig. 8e–h), which was consistent with the prognostic results of single-factor calculations that included only PRR11 expression.
Associations between the PRR11 expression and immune infiltration level in BLCA from the ESTIMATE algorithm. (a–d) The expression of PRR11 has a significant negative correlation with the immunescore and a positive correlation with tumorpurity score of BLCA samples based on the ESTIMATE algorithm in the TCGA-BLCA and GSE48276 dataset. The horizontal axis in the figure represents gene expression, and the vertical axis is purity/immunescore. The density curve on the right represents the distribution trend of purity/immunescore; the upper density curve represents the distribution trend of genes. (e–h) Kaplan–Meier survival curves were used to analyze the correlation between purity/immunescore and OS of BLCA patients. Patients with higher immunescore and lower tumorpurity score have higher OS time in the TCGA-BLCA and GSE48276 dataset.
To gain a deeper understanding of the potential association between PRR11 and immune infiltration, we also analyzed the correlation between the expression of PRR11 and immune marker sets of various immune cells in BLCA using the GEPIA database. Interestingly, the data demonstrated a significant correlation between PRR11 and a large number of immune cell-related molecular markers, such as marker sets of monocyte, neutrophil, natural killer cell , and CD8 + T cell (Table 3). In particular, it should be pointed out that these main roles in regulating tumor immune microenvironment, such as marker sets related to M2 macrophage, tumor-associated macrophages (TAM), Treg and T cell exhaustion, displayed a strong and significant positive correlation with PRR11 (Table 3, Fig. 9a–d). Therefore, we hypothesize that PRR11 indirectly affects the negative regulation of the tumor immune microenvironment and influences the phenotype of BLCA by regulating immune cell infiltration.
Discussion
Bladder cancer is one of the three major malignant tumors of the genitourinary system. However, the current diagnosis and treatment methods for this disease are limited, so exploring novel tumor markers and therapeutic targets is a significant direction in this field. PRR11 is a relatively new potential candidate oncogene, which may play an important role in the initiation and progression of cancer. Several studies have reported that PRR11 overexpression promotes the proliferation, migration, and invasion of ovarian cancer cells by activating the PI3K/AKT/β-Catenin pathway24, and it also can promote the progression of breast cancer by activating EMT39. In addition, PRR11 silencing not only causes cell cycle arrest, inhibits colony formation, reduces cell proliferation, and inhibits the tumorigenic potential of lung cancer cells40, but also inhibits the growth and EMT of liver cancer cells through β-catenin signaling20. PRR11 is also considered a potential new target for the diagnosis and treatment of lung cancer41.
Transcriptome analysis of clinical samples from databases such as TCGA and Oncomine demonstrated that the level of PRR11 mRNA in BLCA was significantly higher than that in non-cancerous tissues, regardless of whether it was observed from the overall sample or the matched sample of a single patient. By analyzing the HPA database, we confirmed the high expression status of PRR11 in BLCA patients at the protein level. In addition, we found that the expression level of PRR11 significantly affected the pathological staging of tumors. Moreover, the multivariate analysis revealed that PRR11 expression was an independent prognostic factor of BLCA patients. Therefore, our findings suggest that PRR11 is up-regulated in the vast majority of BLCA samples and is involved in the pathological process of BLCA tissues. As a potential prognostic marker, PRR11 deserves more detailed clinical verification.
The analysis using the LinkedOmics database revealed that most genes co-expressed with PRR11 in BLCA exhibited prognostic trends, and six genes were statistically significant in BLCA patients. Moreover, these co-expressed genes were highly concentrated in the cell cycle, DNA repair and replication, cancer-related microRNA, ErbB signaling pathway, and other tumor-related biological pathways, consistent with the known function of PRR11. Future research on PRR11 in BLCA may reveal the relevant molecular mechanism.
TMB is a promising biomarker for cancer prognosis that can direct immunotherapy in the era of precision medicine44. Previous studies have demonstrated that TMB can be used as a biomarker to improve immunotherapy45. In addition, MSI is an important biomarker for immune checkpoint inhibitors (ICI)46. According to our research, the PRR11 expression in BLCA positively correlates with TMB but does not significantly correlate with MSI. This may indicate that the expression level of PRR11 affects the TMB of BLCA, thereby influencing the patient's response to immune checkpoint suppression therapy. Additionally, we observed a positive correlation between PRR11 expression and MMR gene expression. Therefore, patients with high PRR11 expression and TMB levels may have better prognoses following ICI treatment in BLCA.
It is well known that the immune environment constructed by multiple immune cells as part of TME has been considered critical to the progression of tumors and the overall effectiveness of cancer treatments, including chemotherapy and radiotherapy, especially immunotherapy13,14,42,43. According to the calculation results of the ESTIMATE algorithm, PRR11 expression was significantly correlated with immune score and tumor purity scores in BLCA samples, and patients grouped by immune score or tumor purity scores also had a constant prognostic relationship with patients grouped with PRR11 expression. These findings clearly show that increasing the level of immune cell infiltration and decreasing the expression of PRR11 may improve the tumor immune microenvironment and prognosis of BLCA patients.
The TIMER database analysis visually revealed that the expression of PRR11 is correlated with the immune cell infiltration in BLCA. In addition, the results from the TISIDB database and GEPIA database demonstrated that the PRR11 in BLCA was positively correlated with the Th2, CD8 + T cells, M2 macrophages, TAM, Treg, NK, neutrophils, monocytes, and the markers of T cell exhaustion, such as PDCD1, TIM3, CTLA4, and LAG3.
Macrophages in the tumor immune microenvironment tend to become TAM, which is differentiated from monocytes migrating to tumor stroma to drive tumor progression, invasion, and metastasis47,48. The relationship between PRR11 and monocytes is supported by TAM data in our findings. In addition, reports indicate that Th1 cytokines activate M1 macrophages to exert anti-tumor effects, whereas Th2 cytokines can activate M2 macrophages to exert tumor-promoting effect49. Although CD4 + T cells and PRR11 did not show a significant correlation in our analysis results, we speculate that PRR11 in BLCA may not affect the total CD4 + T cell infiltration level, but it can regulate the subtypes of infiltrating immune cells. This means that PRR11 increases the proportion of Th2 cells to produce an immunosuppressive TME. However, considering that the correlation between PRR11 and Th2-related molecular markers was insignificant, we believe there may be other indirect control methods. The literature illustrates that tumor-infiltrating dendritic cells also tend to promote immune suppression and tolerance rather than driving anti-tumor immunity50, and some dendritic cells are derived from monocytes (moDCs)51. Neutrophils in tumor tissues typically transform into N2 subtypes with pro-tumor effects50.
Tregs promote tumor growth and expansion by supporting the host's immune response, accelerating angiogenesis, and tissue remodeling52. Especially functional FOXP3+ Treg cells can inhibit cytotoxic CD8+ T cells from attacking tumor cells and promote tumor development53.
Finally, it should be pointed out that CD8+ T cells are an independent prognostic factor in certain cancers, such as pancreatic cancer, and they can play a tumor suppressor role in the tumor immune microenvironment54,55,56,57. Although there was a significantly positive correlation between PRR11 and CD8+ T cells in BLCA, it could not be ignored that it also demonstrated a broad positive correlation with the four molecular markers of T cell exhaustion (PDCD1, CTLA4, LAG3, TIM3). Based on this, we believe that PRR11 may increase the level of CD8+ T cell infiltration in BLCA tumor tissues to a certain extent, but these tumor-infiltrating CD8 + T cells cannot exert their ability of tumor suppressor due to T cell exhaustion and the suppressive immune microenvironment. These findings also highlight the complex mechanism of PRR11 in regulating the tumor immune microenvironment.
Therefore, we suggest that PRR11 in BLCA tends to induce a tumor-suppressive immune microenvironment, and the up-regulation of its expression may directly or indirectly alter the type and level of immune cell infiltration. In this way, the tumor immune microenvironment of BLCA patients is altered to a tumor-promoting type, which has a negative impact on patient prognosis.
In conclusion, PRR11 has the potential as a molecular marker for the poor prognosis of BLCA. Increased expression of PRR11 is associated with deterioration of clinical characteristics, such as tumor pathological grade and prognosis (OS and DFS). In addition, the expression of PRR11 in BLCA is significantly related to tumor immune infiltration, which will inevitably have a profound effect on the prognosis of BLCA. Moreover, given the relationship between PRR11 and the TMB and MMR genes, it is reasonable to assume that PRR11 has considerable value in ICI. Therefore, in the future, we can try to evaluate the prognosis and risk stratification of patients by detecting the expression level of PRR11 in surgical specimens from BLCA patients, as well as develop immunotherapy drugs targeting PRR11 based on the immune microenvironment.
Data availability
All relevant data are within the paper.
Abbreviations
- PRR11:
-
Proline-rich protein
- BLCA:
-
Bladder urothelial carcinoma
- TCGA:
-
The Cancer Genome Atlas
- GEO:
-
Gene Expression Omnibus
- TMB:
-
Tumor mutational burden
- MSI:
-
Microsatellite instability
- MMR:
-
Mismatch repair
- MLH1:
-
MutL homologous gene
- MSH:
-
MutS homologous gene
- PMS2:
-
Postmeiotic segregation increased 2
- EPCAM:
-
Epithelial cell adhesionmolecule
- TURB:
-
Transurethral resection
- TME:
-
Tumor microenvironment
- EMT:
-
Epithelial-mesenchymal transition
- FPKM:
-
Fragments per kilobase per million
- GTEx:
-
Genotype-Tissue Expression
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under curve
- HPA:
-
Human Protein Atlas
- GEPIA:
-
Gene expression profiling interactive analysis
- OS:
-
Total survival
- TIMER:
-
Tumor IMmune Estimation Resource
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GSEA:
-
Gene set enrichment analysis
- RFS:
-
Relapse-free survival
- TAM:
-
Tumor-associated macrophages
- moDCs:
-
Dendritic cells are derived from monocytes
References
Ferlay, J. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359 (2015).
Freedman, N. D., Silverman, D. T., Hollenbeck, A. R., Schatzkin, A. & Abnet, C. C. Association between smoking and risk of bladder cancer among men and women. JAMA 306, 737 (2011).
Lai, M. N., Wang, S. M., Chen, P. C., Chen, Y. Y. & Wang, J. D. Population-based case-control study of Chinese herbal products containing aristolochic acid and urinary tract cancer risk. J. Natl. Cancer Inst. 102, 179 (2010).
Chen, C. J., Chuang, Y. C., You, S. L., Lin, T. M. & Wu, H. Y. A retrospective study on malignant neoplasms of bladder, lung and liver in blackfoot disease endemic area in Taiwan. Br. J. Cancer 53, 399 (1986).
Tsai, S. M., Wang, T. N. & Ko, Y. C. Cancer mortality trends in a blackfoot disease endemic community of Taiwan following water source replacement. J. Toxicol. Environ. Health A. 55, 389–404 (1998).
Harling, M., Schablon, A., Schedlbauer, G., Dulon, M. & Nienhaus, A. Bladder cancer among hairdressers: A meta-analysis. Occup. Environ. Med. 67, 351 (2010).
Zeegers, M. P., Swaen, G. M., Kant, I., Goldbohm, R. A. & van den Brandt, P. A. Occupational risk factors for male bladder cancer: results from a population based case cohort study in the Netherlands. Occup. Environ. Med. 58, 590 (2001).
Siegel, R., Ma, J., Zou, Z. & Jemal, A. Cancer statistics, 2014. CA Cancer J. Clin. 64(1), 9–29 (2014).
Mitra, A. P. & Daneshmand, S. Molecular prognostication in bladder cancer. Cancer Treat Res. 175, 165 (2018).
Burger, M. et al. Epidemiology and risk factors of urothelial bladder cancer. Eur. Urol. 63, 234 (2013).
Sylvester, R. J. et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: A combined analysis of 2596 patients from seven EORTC trials. Eur. Urol. 49, 465–475 (2006).
Baumgart, S. et al. MicroRNAs in tumor samples and urinary extracellular vesicles as a putative diagnostic tool for muscle-invasive bladder cancer. J. Cancer Res. Clin. Oncol. 145, 2725 (2019).
Pitt, J. M. et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 27, 1482 (2016).
Kalinski, P. & Talmadge, J. E. Tumor immuno-environment in cancer progression and therapy. Adv. Exp. Med. Biol. 1036, 1 (2017).
Wang, M. et al. Role of tumor microenvironment in tumorigenesis. J. Cancer 8, 761 (2017).
Cao, R., Yuan, L., Ma, B., Wang, G. & Tian, Y. Tumour microenvironment (TME) characterization identified prognosis and immunotherapy response in muscle-invasive bladder cancer (MIBC). Cancer Immunol. Immunother. 70, 1 (2021).
Sjodahl, G. et al. Infiltration of CD3(+) and CD68(+) cells in bladder cancer is subtype specific and affects the outcome of patients with muscle-invasive tumors. Urol. Oncol. 32, 791 (2014).
Hu, B. et al. Blockade of DC-SIGN(+) tumor-associated macrophages reactivates antitumor immunity and improves immunotherapy in muscle-invasive bladder cancer. Cancer Res. 80, 1707 (2020).
Cao, J. et al. Screening and identifying immune-related cells and genes in the tumor microenvironment of bladder urothelial carcinoma: Based on TCGA database and bioinformatics. Front. Oncol. 9, 1533 (2019).
Qiao, W., Wang, H., Zhang, X. & Luo, K. Proline-rich protein 11 silencing inhibits hepatocellular carcinoma growth and epithelial-mesenchymal transition through beta-catenin signaling. Gene 681, 7 (2019).
Zhang, H. et al. PRR11 promotes cell proliferation by regulating PTTG1 through interacting with E2F1 transcription factor in pan-cancer. Front. Mol. Biosci. 9, 877320 (2022).
Wang, Y. et al. PRR11 and SKA2 gene pair is overexpressed and regulated by p53 in breast cancer. BMB Rep. 52, 157 (2019).
Zhang, L. et al. Silencing of PRR11 suppresses cell proliferation and induces autophagy in NSCLC cells. Genes Dis. 5, 158 (2018).
Zhu, J., Hu, H., Wang, J., Yang, Y. & Yi, P. PRR11 overexpression facilitates ovarian carcinoma cell proliferation, migration, and invasion through activation of the PI3K/AKT/beta-catenin pathway. Cell Physiol. Biochem. 49, 696 (2018).
Ma, H., Yang, W., Wang, X. & Dai, G. PRR11 promotes proliferation and migration of colorectal cancer through activating the EGFR/ERK/AKT pathway via increasing CTHRC1. Ann. Clin. Lab. Sci. 52, 86–94 (2022).
Olsson, H. S. et al. PRR11 unveiled as a top candidate biomarker within the RBM3-regulated transcriptome in pancreatic cancer. J. Pathol. Clin. Res. 8, 65–77 (2022).
Zhou, F. et al. Proline-rich protein 11 regulates epithelial-to-mesenchymal transition to promote breast cancer cell invasion. Int. J. Clin. Exp. Pathol. 7, 8692 (2014).
Lin, J. et al. A robust 11-genes prognostic model can predict overall survival in bladder cancer patients based on five cohorts. Cancer Cell Int. 20, 402 (2020).
Vivian, J. et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 35, 314 (2017).
Rhodes, D. R. et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9, 166 (2007).
Tang, Z. et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucl. Acids Res. 45, W98 (2017).
Siemers, N. O. et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS ONE 12, e179726 (2017).
Li, T. et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, e108 (2017).
Ru, B. et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 35, 4200 (2019).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013).
Vasaikar, S. V., Straub, P., Wang, J. & Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucl. Acids Res. 46, D956 (2018).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 28, 27–30 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Prot. Sci. 28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucl. Acids Res. 49, D545–D551 (2021).
Ji, Y. et al. PRR11 is a novel gene implicated in cell cycle progression and lung cancer. Int. J. Biochem. Cell Biol. 45, 645 (2013).
Zhang, C. et al. PRR11 regulates late-S to G2/M phase progression and induces premature chromatin condensation (PCC). Biochem. Biophys. Res. Commun. 458, 501 (2015).
Oliver, A. J. et al. Tissue-dependent tumor microenvironments and their impact on immunotherapy responses. Front. Immunol. 9, 70 (2018).
Roma-Rodrigues, C., Mendes, R., Baptista, P. V. & Fernandes, A. R. Targeting tumor microenvironment for cancer therapy. Int. J. Mol. Sci. 20, 1 (2019).
Steuer, C. E. & Ramalingam, S. S. Tumor mutation burden: Leading immunotherapy to the era of precision medicine?. J. Clin. Oncol. 36, 631 (2018).
Devarakonda, S. et al. Tumor mutation burden as a biomarker in resected non-small-cell lung cancer. J. Clin. Oncol. 36, 2995 (2018).
Boland, C. R. & Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 138, 2073 (2010).
Ngambenjawong, C., Gustafson, H. H. & Pun, S. H. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv. Drug Deliv. Rev. 114, 206 (2017).
Allavena, P., Sica, A., Garlanda, C. & Mantovani, A. The Yin-Yang of tumor-associated macrophages in neoplastic progression and immune surveillance. Immunol. Rev. 222, 155 (2008).
den Breems, N. Y. & Eftimie, R. The re-polarisation of M2 and M1 macrophages and its role on cancer outcomes. J. Theor. Biol. 390, 23 (2016).
Fukunaga, A. et al. CD8+ tumor-infiltrating lymphocytes together with CD4+ tumor-infiltrating lymphocytes and dendritic cells improve the prognosis of patients with pancreatic adenocarcinoma. Pancreas 28, e26 (2004).
Yang, Z. J. et al. Functions of dendritic cells and its association with intestinal diseases. Cells-Basel. 10, 1 (2021).
Hiraoka, N., Onozato, K., Kosuge, T. & Hirohashi, S. Prevalence of FOXP3+ regulatory T cells increases during the progression of pancreatic ductal adenocarcinoma and its premalignant lesions. Clin Cancer Res. 12, 5423 (2006).
Facciabene, A., Motz, G. T. & Coukos, G. T-regulatory cells: Key players in tumor immune escape and angiogenesis. Cancer Res. 72, 2162 (2012).
Yuan, C. H. et al. Amphiregulin activates regulatory T lymphocytes and suppresses CD8+ T cell-mediated anti-tumor response in hepatocellular carcinoma cells. Oncotarget 6, 32138 (2015).
Bhattacharya, N. et al. Normalizing microbiota-induced retinoic acid deficiency stimulates protective CD8(+) T cell-mediated immunity in colorectal cancer. Immunity 45, 641 (2016).
Ye, J. et al. Oxymatrine and cisplatin synergistically enhance anti-tumor immunity of CD8(+) T cells in non-small cell lung cancer. FRONT ONCOL. 8, 631 (2018).
Ino, Y. et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer. 108, 914 (2013).
Acknowledgements
Here we express our sincere respect to the founders and data collectors of TCGA, GEO, Oncomine, GEPIA, HPA, LinkedOmics, TIMER, ESTIMATE, TISIDB, KEGG and other online databases. It is precisely because of your selfless dedication and exquisite skills that biomedicine can have such a broad platform for clinical researchers like us.
Funding
This research was funded by the Social Welfare and Basic Research Project Fund of Zhongshan City (2021B1088).
Author information
Authors and Affiliations
Contributions
W.N. designed and supervised the study, L.Y., C.Y., W.N., J.Z., and Q.W. collected and analyzed data, W.N., X.D. and M.C. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ni, W., Yi, L., Dong, X. et al. PRR11 is a prognostic biomarker and correlates with immune infiltrates in bladder urothelial carcinoma. Sci Rep 13, 2051 (2023). https://doi.org/10.1038/s41598-023-29316-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29316-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.