Abstract
The characteristics of long-term rice straw decomposition and succession in the bacterial community in the double-rice system are still unclear. Here a 2-year continuous straw bag decomposition experiment was conducted to explore changes in nutrient release, enzyme activity, and bacterial community composition during rice straw decomposition in the double-rice system in Southeast China. After burial in soil, the cumulative dry matter loss rates of rice straw were 38.9%, 72.6%, and 82.7% after 2, 12, and 24 months, respectively. The change in the release rate of straw nitrogen and phosphorus was similar to the dry matter loss, but 93.5% of straw potassium was released after 1st month. Bacterial abundance and community diversity in straw increased rapidly, reaching peaks after 7 and 12 months, respectively. Straw extracellular enzyme activities were the highest in the first 2 months and then gradually decreased over time, and they significantly and positively correlated with straw decomposition rate. Straw decomposition was dominated by copiotrophic Bacilli and Flavobacteriia in the early stages and evolved to be dominated by oligotrophic Acidobacteria, Anaerolineae, Deltaproteobacteria, Saccharibacteria, and Sphingobacteriia in the later stages. Changes in the C/N and K content of straw are the main reasons for bacterial community succession during rice straw decomposition. This study can provide a scientific basis for developing efficient decomposing bacteria agents for rice straw.
Similar content being viewed by others
Introduction
Crop straw, as an organic by-product of agricultural production, contains rich nutrients needed for plant growth. It has been estimated that 31.6%, 22.5% and 28.8% of the amount of nitrogen required for rice, wheat and corn production can be supplied by returning all the corresponding crop straw to the field in China1. The amount of potassium that can be supplied by returning rice, wheat and corn straw to the field in the current season is 4.99, 1.93 and 4.79 million tons, respectively2. After crop straws are returned to the soil, their mineral nutrients are gradually released during the decomposition process, and the process is mainly regulated by soil microbes. Straw quality and other environmental conditions, such as soil water and temperature, also affect the decomposition process by influencing the abundance and community composition of soil microbes3,4,5. Therefore, exploring the nutrient release pattern during straw decomposition can guide the fertilization measures after returning rice straw to field.
Microorganisms and enzymes play an important role in straw decomposition. Previous studies showed that the rapid decomposition of soluble organic components such as soluble sugars, amino acids and proteins, as well as hemicellulose and cellulose, occurs within the straw due to soil microbial activity in the early stages of straw decomposition6. At this stage, copiotrophic flora such as Actinobacteria, Firmicutes, and Proteobacteria dominated in straw decomposition7,8, and β-N-acetylaminoglucosidase and leucine aminopeptidase also showed high activity and gradually decreased with the consumption of easily degradable substrates9. Difficult-to-decompose substances in straw (e.g., lignin, waxes, and tannins) gradually accumulated and were slowly decomposed by specific microorganisms in the late stages of straw decomposition10,11. At this stage, the abundance of oligotrophic Acidobacteria, Chloroflexi, and Saccharibacteria gradually increased12,13, and the activities of β-cellobiohydrolase and phenol oxidas showed high activity14,15. The decomposition of crop straw is a long-term process. Most of the existing studies are short-term decomposition experiments, with the majority of studies on decomposition of crop straw in the growing season16,17. During the non-growing season, the conversion of organic and inorganic components of straw into soil is still ongoing, and the composition of microbial communities is also changing. However, there is still a lack of research on characteristics of long-term straw decomposition and microbial community succession18,19.
Rice is a major grain crop in south China and produces more than 200 million tons of straw every year1. The input of straw nutrient resources has great potential for reducing fertilizer application. Sufficient soil water and higher temperature are beneficial to the straw decomposition and are not the limiting factors in the paddy soil20, and the rice straw decomposition may be mainly affected by biological factors. Therefore, it is of great significance to study the change in microbial abundance and community composition during the straw decomposition process to reveal the microbial mechanism of straw decomposition. Previous studies found that differences in crop straw components also lead to differences in characteristics of straw decomposition and microbial community succession21. Moreover, most of the previous experiments on straw decomposition in the field were conducted during the growing season of the crop17, and the characteristics of rice straw decomposition and the structure of the microbial community in long-term straw decomposition in the double-rice system are still unclear. Given this, A 2-year continuous experiment on rice straw decomposition was established in a double-cropping rice system in southeastern China. Our objectives were to (i) determine the dynamic of nutrient release and enzyme activity changes in straw and (ii) clarify the characteristics of succession in bacterial community composition and the factors influencing the succession during the straw decomposition.
Results
Changes in straw dry matter and nutrient release
The cumulative loss rate of straw dry matter mass was 33.7%, 38.9%, 48.1%, 53.9%, 69.0%, 72.6%, 77.6%, and 82.7% after 1, 2, 5, 7, 10, 12, 20, and 24 months since straw burial in soil (Fig. 1). The changes in the release rates of N and P from straw across the decomposition process were similar to the changes in dry matter loss rates, and they met the fitting formula of y = 0.16x2 − 5.97x + 81.6 (x = time, y = release rate of nutrients or dry matter from the straw, R2 = 0.78). 93.5% of the total straw K was released after 1 month. The straw C/N increased in the initial 2 months and gradually decreased with prolonged experimental time.
Changes in straw enzyme activity and bacterial abundance
The activities of BG, CB, XYL, NAG, and LAP were the highest in the initial stage of straw decomposition and then decreased significantly with test time. The changes in the activities of CB, NAG, and LAP with the straw test time can be explained by the fitting formula ‘y = 4.66x2 − 163.92x + 1474.1’ (x = time, y = enzyme activity, R2 = 0.87) (Fig. 2a). The copy number of bacterial 16S rRNA gene of straw gradually increased in the first 7 months after the straw was buried in soil, and then decreased with experimental enlarging time (Fig. 2b).
Changes in enzyme activity (a) and bacterial abundance (b) during straw decomposition. Different lower letters at the top of the columns present significant differences among decomposition periods (P < 0.05). BG, CB, XYL, NAG, and LAP are β-glucosidase, β-cellobiohydrolase, β-xylosidase, β-N-acetylglucosaminidase, and Leucine aminopeptidase activities, respectively.
The bacterial α-diversity during straw decomposition
The OTUs, ACE, Chao1 and Shannon index of straw bacteria increased significantly compared with those in the original straw (before burial in soil) and showed a trend of first increasing and then stabilizing with the passage of test time, and the maximum was all in the 12th month, but the Simpson index shows the opposite trend to the Shannon index (Table 1).
The bacterial β-diversity and influence of environmental factors on community composition during straw decomposition
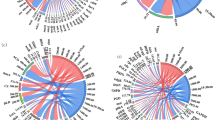
Principal component analysis showed that the bacterial phylum community composition in the 0 and 2nd months was significantly separated from other periods along the PC1 axis (Fig. 3a). Starting from the 0 month, the composition of the bacterial community is roughly distributed along the PC1 axis from the positive axis to the negative axis, and the community composition at 7–24 months did not separate significantly. The bacterial community in the initial straw was dominated by Actinobacteria and Proteobacteria, and Bacteroidetes and Firmicutes dominated the community in the 2nd month. The leading roles of Acidobacteria, Chloroflexi, Ignavibacteriae, Gracilibacteria, Parcubacteria, Saccharibacteria, Spirochaetae, and Verrucomicrobia increased significantly during the 7–24 months. For bacteria at the genus level, the bacterial community in the initial straw is dominated by Actinobacteria and Gammaproteobacteria; Bacilli and Flavobacteriia dominate the community composition in the 2nd month; Acidobacteria, Anaerolineae, Deltaproteobacteria, Saccharibacteria, and Sphingobacteriia dominated the community composition during the 7–24 months of straw burial (Fig. 3b).
Redundancy analysis showed that bacterial community composition at the phylum level was mainly influenced by straw K and C/N, while bacterial community composition was mainly influenced by straw C/N, K and P at the class level (Fig. 4a,b).
Succession of bacterial community composition during straw decomposition
The relative abundance of phylum Proteobacteria during straw decomposition was the highest, accounting for 39.9‒52.0% of the total bacterial abundance of the sample, followed by Bacteroidetes (2.3‒30.3%), Actinobacteria (2.6‒8.7), Chloroflexi (0.1‒13.3%), Acidobacteria (0.2‒16.5%), Saccharibacteria (0.1‒4.4%), Firmicutes (1.0‒12.7%), Verrucomicrobia (0.7‒2.0%), Gracilibacteria (0.01‒1.5%), Parcubacteria (0.01‒2.9%), Spirochaetae (0.01‒3.7%) and Ignavibacteriae (0.01‒2.4%) (Fig. 5a). After the straw was buried in the soil, the relative abundance of Proteobacteria and Actinobacteria decreased gradually, and Acidobacteria, Chloroflexi and Saccharibacteriamen gradually increased with increasing experimental time, while Bacteroides and Firmicutes presented an increase–decrease change across the experiment process, and the highest abundance occurred in the 2nd month (Fig. S1a).
The bacterial class Alphaproteobacteria has the highest abundance, accounting for 15.4% to 27.7% of all components of the sample, followed by Deltaproteobacteria (1.3‒16.1%), Acidobacteria (0.2‒16.5%), Betaproteobacteria (3.1‒13.6%), Gammaproteobacteria (4.4‒9.3%), Actinobacteria (2.6‒10.9%), Flavobacteriia (0.01‒20.9%), Anaerolineae (0.01‒6.9%), Bacilli (0.1‒12.2%), Sphingobacteriia (1.2‒7.8%) and Saccharibacteria (0.08‒4.4%) (Fig. 5b). The abundance of Acidobacteria and Saccharibacteria gradually increased with the passage of test time, while the relative abundance of Flavobacteria and Gammaproteobacteria gradually decreased. The relative abundance of Bacilli, Alphaproteobacteria, Betaproteobacteria, and Sphingobacteriia showed an increasing–decreasing trend, which was the highest in the 2nd month. The relative abundance of Anaerolineae and Deltaproteobacteria first increased and then decreased, reaching the peak at the 12th and 20th months, respectively (Fig. S1b).
Discussion
Climate condition is considered to be the decisive factor for the decomposition of organic materials22,23. In this study, the temperature (approximately 20 ℃) in the early stage of straw decomposition provided favorable conditions for the rapid decomposition of straw. The decomposition rate of rice straw in the early stage (the first 3 months) was higher than that in the late stage. Because the labile fractions were abundant and easily decomposed in the early stage, the decomposition rate of straw gradually slowed down with the accumulation of recalcitrant components in the later stage24,25. Moreover, straw decomposition rate showed significant differences at similar conditions (similar temperature and moisture) between the two experimental years, and the decomposition was slower at the later stage relative to that at the early stage, although the high bacterial abundance at the later stage in this study, suggesting that in paddy fields of Southeast China, the straw chemical composition is a more important factor influencing its decomposition compared to external environmental conditions.
The release rate of straw N and P was similar to the dry matter loss rate with the increasing test month (Fig. 1). Previous studies have shown that about 40% of the P in the straw is involved in the formation of the cell wall, and N mainly exists in the straw in the organic state with a high degree of cementation, which leads to a strong synchronization between the release of straw N and P and the loss of straw dry matter16,18. In this study, the release rate of straw K was higher than that of N and P in this study because K in straw can be directly released mainly in the form of K salt26. The N release rate of straw in the first two months was greater than the loss rate of dry matter (C), which may be the main reason for the increase in C/N at the early stages of straw decomposition.
The extracellular enzymes secreted by microorganisms are catalytic agents for straw decomposition, and it is beneficial to clarify the microbial driving mechanism in straw decomposition by analyzing their changes in the process of straw decomposition27,28. The activities of the five extracellular enzymes involved in the conversion of C and N were significantly higher in the early stage of decomposition (Fig. 2). This may be that sufficient straw C and N in the early stage promoted the growth and reproduction of microorganisms involved in the C and N cycles, and a large number of related microorganisms further induced an increase in the secretion of C and N cycle enzymes29. In addition, it was shown that there was a significant correlation between enzyme activity magnitude during straw decomposition and straw C/N ratio (P < 0.05), which further indicates that the enzyme activity secreted by microorganisms is strongly influenced by the straw C and N content (Fig. S1). In this study, β-glucosidase still maintained a higher enzyme activity in the late stage of decomposition, indicating that it plays a very important role in the late stage of straw decomposition, which is consistent with the results of previous studies on the changes of the enzyme activity of recalcitrant organic matter decomposition14. Microorganisms are the main decomposers of straw, and their abundance and community composition can affect the rate of straw decomposition25. In this study, bacterial abundance and α-diversity continued to increase when the rate of straw decomposition decreased, which may be caused by a decrease in bacterial abundance that lags behind the decrease in residue biomass26,30, and we suggest that as straw decomposers, soil microorganisms near the straw bag continue to colonize the straw after obtaining a large amount of carbon source, which is the main reason why bacterial α-diversity continues to increase even after the reduction of straw decomposition rate.
This study showed that the copiotrophic phylum Bacteroidetes, Firmicutes, and Proteobacteria dominated bacterial community composition at the initial stage of decomposition (Fig. 3), which is because the labile C fractions and other easily degradable components in the early stage of straw decomposition provide an easily accessible and rich source of energy for the growth and reproduction of these bacteria, thus promoting the increase of their populations8,12. The Acidobacteria, Chloroflexi, and Saccharibacteria gradually increased with the progress of the experiment, these three phyla were oligotrophic populations and dominated during the decomposition of residues with high recalcitrant fractions (e.g. lignin, cellulose) and poor nutrients in the later stage of residue decomposition13. Bacilli (soil saprophyte) was mainly found in the early stage of straw decomposition and consistent with the previous results31. The study found that the abundance of Actinobacteria increased in the late stage. Bernard et al. also reported the increase of Actinobacteria in the final stage of wheat straw degradation and believed that these oligotrophic bacteria might participate in the stimulating effect of straw decomposition32. The correlation analysis between the straw decomposition rate at different sampling periods and the relative abundance of different taxa showed that the rate of straw decomposition was significantly and positively correlated with the relative abundance of Proteobacteria, Bacteroidetes, Alphaproteobacteria, Bacilli, Betaproteobacteria, Gammaproteobacteria, Sphingobacteriia, and Flavobacteriia (Fig. S1), indicating that increasing the abundance of these taxa may facilitate the increase of straw decomposition rates. In addition, we also found that the changes in the relative abundance of different classes of bacteria within the same phylum level were inconsistent throughout the experiment (Fig.S2), implying that bacterial communities within the same phylum have functional differences at different stages of straw decomposition13.
The microorganisms colonizing crop residues mainly come from the soil near the residues24,33. However, the effect of soil biochemical properties near the straw bags on them was not explored in this experiment, which should be taken into account in future studies. In addition, fungi can effectively break down recalcitrant compounds within the straw by secreting residue degrading enzymes15,34. In future studies, we will explore the synergistic effects of fungal and bacterial communities during the decomposition of rice straw, which in turn will provide a reference for the isolation of microbial strains for rice straw degradation.
Conclusions
The rate of rice straw decomposition and nutrient release was greater at the first 2 months after burial in soils, and gradually decreased with increasing experimental time in Southeast China. Rice straw decomposition rate showed a significant positive correlation with extracellular enzyme activity. Classes Bacilli and Flavobacteriia played a key role in the early stage of straw decomposition, while Acidobacteria, Anaerolineae, Deltaproteobacteria, Saccharibacteria, and Sphingobacteria dominated bacterial communities in the later stage. The change in straw chemical composition was the main factor affecting straw bacterial flora in Southeast China.
Material and methods
Experimental site
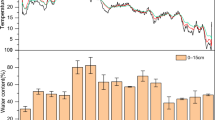
The straw decomposition experience was carried out at Nanchang Experimental Station of Jiangxi Academy of Agricultural Sciences (116°20'E, 28°15'N) from October 2014 to October 2016. The area has a subtropical monsoon climate with double-cropping rice as the main planting system. The annual average temperature and precipitation are 17.7 °C and 1374 mm, respectively. The physicochemical properties of the original soil (0–20 cm) before the start of the experiment: pH 6.6 (water: soil = 2.5:1), organic carbon 9.2 g kg−1, total nitrogen (N) 1.31 g kg−1, mineral N (nitrate N + ammonium N) 57.0 mg kg−1, available phosphorus (P) 13.6 mg kg−1, available potassium (K) 108.0 mg kg−1, the data of rainfall and temperature during the testing period were shown in Fig. 6.
Experimental design
Rice straws (stems and leaves) were collected from the experimental field during the late rice harvest in October 2014. The straws were dried at 65 °C and cut into pieces (1–2 cm × 0.3–1 cm), and 12 g straw (8 t hm−2) was put into a nylon net bag (15 cm × 10 cm). Subsequently, 60 straw bags were embedded in the paddy soil (15 cm depth, 20 cm spacing distance). In order to avoid the influence of fertilizer on the determination of residual nutrients, no fertilizer was applied to the field during the experiment.
Straw bag sampling
Six straw bags were collected at 0, 1, 2, 5, 7, 10, 12, 20, and 24 months after being buried, the soil particles and soil animals attached to the straw bags were removed, and the samples were transported to the laboratory immediately in the ice box. After three straw bags were dried to a constant weight at 65 ℃, the dry matter and nutrient content were measured. The other three straw bags at 0, 2, 7, 12, 20 and 24 months were stored at −70 °C for enzyme activity and bacterial community composition determination.
Straw dry matter, nutrient and enzyme activity analysis
The straw sample was dried at 70 °C to a constant weight and then the dry matter was determined. The C and N of straw were determined by C/N element analyzer (Model CN, Vario Macro Elementar, Germany), P was determined by the H2SO4–H2O2 digestion-vanadium molybdenum yellow colorimetric method, K was measured by atomic absorption spectrophotometer (SpectAA-50/55, Varian, Australia)35. The cumulative loss rate of straw dry matter mass (A) and the release rate of straw nutrients (B) were calculated using the equation as following:
W1 and m1 are the original dry matter weight of straw (g) and the original straw nutrient content (g kg−1); W2 and m2 are the dry matter weight (g) and nutrient content of straw after decomposing at different sampling times (g kg−1).
A fresh sample of straw equivalent to 0.3 g dry weight was weighed and placed in a centrifuge tube. The sample was thoroughly mixed with 50 ml sodium acetate buffer (50 mM) using a vortexer, and then the suspension was poured into a beaker, and a magnetic rotor was put in to maintain a uniform suspension. Buffer, sample suspension, 10 mM control and 200 mM substrate were sequentially added to a 96-well microtiter plate, incubated at 25 °C in the dark for 4 h, and then used with a microplate reader (Scientific Fluoroskan Ascent FL, ThermoFischer Scientific, Waltham, MA) for fluorescence quantification. The measurement conditions were excited at 365 nm, and the signal was recovered at 450 nm, followed by determination of β-glucosidase, β-cellobiohydrolase, β-xylosidase, β-N-acetylglucosaminidase, and Leucine aminopeptidase activities36,37. The enzyme activity unit is nmol g−1 h−1.
Microbial total DNA extraction, PCR amplification, and Illumina NovaSeq sequencing
A 0.2 g straw sample was used for the microbial DNA extraction using the FastDNA® SPIN kit (MP Biomedicals, Illkirch, France). The extracted DNA was purified using Ultra-Clean DNA cation kit (MoBio, Carlsbad, CA, USA) and was checked using 1.0% agarose gel electrophoresis. SYBR Green I was used to quantify the abundance of bacterial 16S rRNA gene copies in the iCycler system (USA) and the results were analyzed by Bio-Rad iQ5 v2.0. The PCR reaction system (20 μl) contains 2 × 10 Super Mix (Bio-Rad, USA), 10 μM primers (515F and 806R) and 1 μl of 1/10 diluted DNA. PCR was performed using the following thermal cycling program: 95 °C for 1 min, followed by 40 cycles at 94 °C for 15 s, 55 °C for 34 s, and 72 °C for 15 s. The plasmid containing bacterial DNA was diluted into different gradients to prepare a reaction standard curve to calculate the bacterial abundance. The V3-V4 region of the bacterial 16SrRNA gene was amplified by PCR using primers 338F (5'-ACTCCTACGGGAGGCAGCAG-3') and 806R (5'-GGACTACHVGGGTWTCTAAT-3') (95 °C for 2 min; 95 °C for 30 s, 56 °C for 30 s, 72 °C for 60 s, 30 cycles; 72 °C for 5 min final extension), the amplified product was recovered with 2% agarose gel, purified with AxyPrep DNA purification kit (Axygen Biosciences, Union City, CA, USA), and quantified with QuantiFluor ™-ST (Promega, USA). The purified PCR products were subjected to high-throughput sequencing using the Illumina MiSeq platform. Then the original sequence is subjected to quality control and processing using QIIME (V1.17). In order to avoid deviations caused by differences in sequencing depth, 33,480 sub-samples of each sample were randomly selected for read leveling and used for bacterial diversity and community composition analysis. UPARSE (V7.1) was used to classify OTUs according to 97% similarity, and UCHIME was used to identify and remove chimeric sequences. The Silva database was used to analyze the classification of each 16S rRNA gene sequence. Shannon, Simpson, Chao1 and ACE indices were calculated by Mothur software. The sequencing results were stored in the NCBI database (Accession number: PRJNA623251).
Statistical analysis
The bacterial gene abundance, diversity index, and relative abundance at different sampling times were compared by one-way ANOVA based on Fisher's least significant difference (P < 0.05) using SPSS 19.0 (SPSS, Inc., Chicago, IL, USA). Canoco (V5.0, Ithaca, NY) was used for principal component analysis and redundancy analysis, and the R package "Vegan" was used for the ADONIS analysis (999 permutations) of bacterial communities during straw decomposition.
Statement
All rice straw samples collected in this study have been licensed. All the rice straw experiments were in compliance with relevant institutional, national, and international guidelines and legislation.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Song, D. L. et al. Nutrient resource quantity of crop straw and its potential of substituting. Plant Nutr. Fert. Sci. 24, 1–21 (2018).
Chai, R. S. et al. Potassium resource quantity of main grain crop straw and potential for straw incorporation to substitute potassium fertilizer in China. Plant Nutr. Fert. Sci. 26, 201–211 (2020).
Grandy, A. S. et al. Soil respiration and litter decomposition responses to nitrogen fertilization rate in no-till corn systems. Agric. Ecosyst. Environ. 179, 35–40 (2013).
Sun, Y., Huang, S. & Yu, X. C. Differences in fertilization impacts on organic carbon content and stability in a paddy and an upland soil in subtropical China. Plant Soil 397, 189–200 (2015).
Yang, H. S. et al. Long-term ditch-buried straw return increases functionality of soil microbial communities. CATENA 202, 105316 (2021).
Preston, C. M. et al. Chemical changes during 6 years of decomposition of 11 litters in some Canadian forest sites. Part 2. 13C Abundance, solid-state 13C NMR spectroscopy and the meaning of “lignin”. Ecosystems 12, 1078–1102 (2009).
Fierer, N., Bradford, M. A. & Jackson, R. B. Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364 (2007).
Fan, F. L. et al. Probing potential microbial coupling of carbon and nitrogen cycling during decomposition of maize straw by 13C-DNA-SIP. Soil Boil Biochem. 70, 12–21 (2014).
Baumann, K., Marschner, P. & Smernik, R. J. Residue chemistry and microbial community structure during decomposition of eucalypt, wheat and vetch residues. Soil Boil Biochem. 41, 1966–1975 (2009).
Heitkamp, F. et al. Implications of input estimation, residue quality and carbon saturation on the predictive power of the Rothamsted carbon model. Geoderma 170, 168–175 (2012).
Wang, X. Y. et al. Succession in bacterial community during maize straw decomposition. Plant Nutr. Fert. Sci. 27, 45–53 (2021).
Ghosh, A. et al. Temperature sensitivity of soil organic carbon decomposition as affected by long-term fertilization under a soybean based cropping system in a sub-tropical Alfisol. Agric. Ecosyst. Environ. 233, 202–213 (2016).
Bastian, F. et al. Impact of wheat straw decomposition on successional patterns of soil microbial community structure. Soil Boil Biochem. 41, 262–275 (2008).
Sinsabaugh, R. L. & Follstad Shah, J. J. F. Ecoenzymatic stoichiometry of recalcitrant organic matter decomposition: the growth rate hypothesis in reverse. Biogeochemistry 102, 31–43 (2011).
Zhao, S. C. et al. Dynamic of fungal community composition during maize residue decomposition process in north-central China. Appl. Soil Ecol. 167, 104057 (2021).
Dai, Z. G. et al. Nutrient release characteristic of different crop straws manure. Trans. Chin. Soc. Agric. Eng. 26, 272–276 (2010).
Zeng, L. et al. Characteristics of decomposition, nutrient release and structure change of wheat straw in a fluvo-aquic soil under different nitrogen application rates. Plant Nutr. Fert. Sci. 26, 1565–1577 (2020).
Wu, J. et al. Decomposition characteristics of rapeseed and wheat straws under different rice cultivations and straw mulching models. Sci. Agric. Sin. 44, 3351–3360 (2011).
Lei, W. Y. et al. Decomposition processes of organic materials and their mechanisms of improving soil fertility in cropland ecosystems. Chin. J. Ecol. 30, 1393–1408 (2022).
Nakajima, M. et al. Modeling aerobic decomposition of rice straw during the off-rice season in an Andisol paddy soil in a cold temperate region of Japan: Effects of soil temperature and moisture. Soil Sci. Plant Nutr. 62, 90–98 (2016).
Zhang, H. et al. Decomposition of plant straws and accompanying variation of microbial communities. Acta Pedol. Sin. 56, 1482–1492 (2019).
Vitousek, P. M., Turner, D. R. & Parton, W. J. Litter decomposition on the Mauna Loa environmental matrix, Hawai’i: Patterns, mechanisms, and models. Ecology 75, 418–429 (1994).
Gregorich, E. G. et al. Litter decay controlled by temperature, not soil properties, affecting future soil carbon. Glob. Chang. Biol. 23, 1725–1734 (2017).
Wang, X. Y. et al. Structural convergence of maize and wheat straw during two-year decomposition under different climate conditions. Environ. Sci. Technol. 46, 7159–7165 (2012).
Kamble, P. N. & Baath, E. Comparison of fungal and bacterial growth after alleviating induced N-limitation in soil. Soil Boil Biochem. 103, 97–105 (2016).
Zhao, S. C. et al. Characteristics of maize residue decomposition and succession in the bacterial community during decomposition in Northeast China. J. Integr. Agric. 20, 2–11 (2021).
Namaghi, H. H., Karami, G. H. & Saadat, S. A study on chemical properties of groundwater and soil in ophiolitic rocks in Firuzabad, east of Shahrood, Iran: With emphasis to heavy metal contamination. Environ. Monit. Assess. 174, 1–4 (2011).
Cenini, V. L. et al. Linkages between extracellular enzyme activities and the carbon and nitrogen content of grassland soils. Soil Biol. Biochem. 96, 198–206 (2016).
Qian, R. X. et al. Effects of maize straw residue amendment on the dynamics of enzyme activities related with soil carbon and nitrogen turnover. Chin. J. Soil Sci 51, 1109–1117 (2020).
Herzog, C. et al. Microbial succession on decomposing root litter in a drought-prone Scots pine forest. ISME J. 13, 2346–2362 (2019).
Lee, C. G. et al. Bacterial populations assimilating carbon from 13C-labeled plant straw in soil: analysis by a DNA-SIP approach. Soil Boil Biochem. 43, 814–822 (2010).
Bernard, L. et al. Dynamics and identification of soil microbial populations actively assimilating carbon from 13C-labelled wheat residue as estimated by DNA- and RNA-SIP techniques. Environ. Microbiol. 9, 752–764 (2007).
Marschner, P., Umar, S. & Baumann, K. The microbial community composition changes rapidly in the early stages of decomposition of wheat residue. Soil Biol. Biochem. 4, 445–451 (2011).
De Boer, W. et al. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29, 795–811 (2005).
Lu, R. K. Methods of Agricultural Chemistry Analysis (China Agricultural Science and Technology Press, 2000).
DeForest, J. L. The influence of time, storage temperature, and substrate age on potential soil enzyme activity in acidic forest soils using MUB-linked substrates and l-DOPA. Soil Boil Biochem. 41, 1180–1186 (2009).
Saiya-Cork, K. R., Sinsabaugh, R. L. & Zak, D. R. The effects of long-term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Boil Biochem. 34, 1309–1315 (2002).
Acknowledgements
This project was supported by the National Key Research & Development Program of China (2018YFD0201001).
Author information
Authors and Affiliations
Contributions
X.W. wrote the manuscript. S.Z. provided experimental condition and provided the project fund. P.H., X.X., S.Q., and S.Z. reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., He, P., Xu, X. et al. Characteristics of rice straw decomposition and bacterial community succession for 2 consecutive years in a paddy field in southeastern China. Sci Rep 12, 20893 (2022). https://doi.org/10.1038/s41598-022-25229-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-25229-8
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.