Abstract
Acanthamoeba polyphaga mimivirus (APMV), a species of amoeba-infecting giant viruses, has recently emerged as human respiratory pathogens. This study aimed to evaluate the presence of Mimivirus in respiratory samples, collected from tuberculosis (TB)-suspected patients. The study was performed on 10,166 clinical respiratory samples from April 2013 to December 2017. Mimivirus was detected using a suicide nested-polymerase chain reaction (PCR) and real-time PCR methods. Of 10,166 TB-suspected patients, 4 (0.04%) were positive for Mimivirus, including Mimivirus-53, Mimivirus-186, Mimivirus-1291, and Mimivirus-1922. Three out of four patients, hospitalized in the intensive care unit (ICU), were mechanically ventilated. All patients had an underlying disease, and the virus was detected in both sputum and bronchoalveolar lavage samples. In conclusion, Mimivirus was isolated from TB-suspected patients in a comprehensive study. The present results, similar to previous reports, showed that Mimiviruses could be related to pneumonia. Further studies in different parts of the world are needed to additional investigate the clinical importance of Mimivirus infection.
Similar content being viewed by others
Introduction
The field of virology was transformed in 2003 with the accidental discovery of giant viruses of amoeba. These viruses were antecedently described in terms of their submicroscopic size, which can be determined by optical microscopy. Several reports have shown that giant viruses of amoebae have structural, genetic, and proteomic complexities, which are comparable to those of bacteria and some small eukaryotes and are not expected among viruses. Up to now, Mimivirus, mollivirus, faustovirus, marseillevirus, and pithovirus, as giant viruses of amoebae, have been increasingly identified in humans1.
Acanthamoeba polyphaga mimivirus (APMV) was introduced in 2005 as the main member of the family Mimiviridae. Since then, approximately 100 new Mimivirus strains have been detected by culturing amoebae from soil, water, and human samples2.
These viruses are intra-amoebal pathogens, which are isolated by culturing amoeba from water samples, collected from cooling towers3,4. They may also be causative agents of pneumonia, similar to other amoeba-associated microorganisms (AAMs). Overall, identification of etiological agents is very important in both community- and hospital-acquired pneumonia, because these infections are the leading cause of morbidity and mortality worldwide5.
In previous studies, to determine whether Mimivirus is a causative agent of pneumonia or not, it was inoculated via intranasal routes in mice, and histopathological evidence of acute pneumonia was found in most animals. The results of these studies suggest that this virus can cause pneumonia under experimental conditions6.
However, there is scarce information about the prevalence of Mimiviruses worldwide to determine the mechanisms of diseases caused by these viruses. Also, the frequency of Mimivirus has not been detected in Iranian patients.
Considering the possible role of Mimivirus as an etiological agent of pathogenic respiratory disease, this study aimed to detect Mimivirus DNA in tuberculosis (TB)-suspected patients, referred to the Department of Mycobacteriology and Pulmonary Research of Pasteur Institute of Iran.
Materials and methods
Study population
From April 2013 to December 2017, a total of 10,166 TB-suspected patients, presenting to Pasteur Institute of Iran, were selected. The current study was performed according to the 1975 Declaration of Helsinki and local regulations. It was also approved by the Ethics Committee of Pasteur Institute of Iran. Written informed consent was obtained from all participants and parents/legal guardians.
Detection of Mimivirus by nested-polymerase chain reaction (PCR) and real-time PCR assays
Viral DNA and RNA were extracted from 10,166 respiratory tract samples, including sputum and bronchoalveolar lavage (BAL) samples, using the High Pure Viral Nucleic Acid Kit and the High Pure Viral RNA Kit (Roche Diagnostics Deutschland GmbH, Mannheim, Germany), according to the manufacturers’ instructions, respectively. Also, bacterial and fungal genomes were isolated using Proba-NK DNA Extraction Kit (DNA-Technology Company, Moscow, Russia) and the QIAamp® DNA Mini Kit (QIAGEN, Carlsbad, USA), following the manufacturer’s instructions, respectively.
Also, a suicide nested-PCR assay was used for the identification of Mimivirus DNA in respiratory samples, as previously described5.
For the real-time PCR assay, two different conserved regions of the Mimivirus genome were targeted, according to a studies by Dare et al., and Saadi et al., as previously described7,8. Because we did not have access to Mimivirus as a positive control, we prepared a plasmid, containing amplicons as the positive control; also, distilled water was used as the negative control.
To confirm the results of PCR assays, an AccuPrep® PCR Purification Kit (Bioneer, South Korea) was used to purify nested-PCR and real-time PCR products. Next, PCR products were sequenced using an ABI automated sequencer (Applied Biosystems, Foster City, CA, USA). MEGA Version 6.0 was used to evaluate the raw sequencing data.
Phylogenetic analysis of Mimivirus based on core genes
For evaluating the evolutionary relationship between our isolated viruses and other families of Mimiviruses, phylogenetic analyses were performed, using nucleocytoplasmic large DNA virus (NCLDV) core genes, including major capsid protein, D5 helicase, family B-DNA polymerase, and VV A18 helicase9.
The PCR method was used for sequencing these genomes, using several overlapping amplicons. Two, five, three, and two fragments covering the near full-length genes of the VV A18 helicase, the family B-DNA polymerase, the D5 helicase, and the major capsid protein, respectively, were amplified by PCR using specific primers from Megavirus LBA111 (JX885207.1) isolate (Supplementary Tables 1–8). After purification with a High Pure PCR Product Purification Kit (Roche Diagnostics Deutschland GmbH, Mannheim, Germany), the purified PCR fragments were sequenced, using an ABI automated sequencer (Applied Biosystems, Foster City, CA, USA). MEGA Version 6.0 was used to evaluate the raw sequencing data.
Results
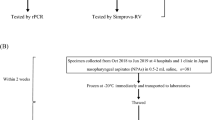
Out of 10,166 TB-suspected patients, four (0.04%) were infected with Mimivirus (Fig. 1). In this study, the best results were obtained using the proposed method by Saadi and colleagues. Also, the result of suicide nested-PCR was positive for all positive samples and negative for the negative controls. The first- and second-round PCR products were 297 bp and 170 bp, respectively, visualized by electrophoresis on 1.5% agarose gel (Supplementary Fig. 1). After sequencing the PCR products that is shown in Supplementary Fig. 2A,B, the phylogenetic tree was drawn. The trees based on the core genes indicated the close relationship of all isolated Mimiviruses with Mimiviruses of lineage C and revealed that these isolates belonged to Mimiviridae clade C (Mimivirus chilensis, Megavirus LBA111, and Courdo11) (Fig. 2).
Neighbor-joining tree constructed based on nucleotide acid sequences of the family B DNA polymerase (A), the D5-ATPase-helicase (B), the major capsid protein (C), and the VV A18 helicase (D) genes. Outgroup was Cafeteria roenbergensis virus. Our isolated Mimiviruses are present within the Mimivirus C lineage.
All positive samples were examined to determine the most common viral, fungal, and bacterial infectious agents of the respiratory tract, that is, Mycobacterium tuberculosis, nontuberculous mycobacteria, Legionella pneumophila, Pseudomonas aeruginosa, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, Haemophilus influenzae, Staphylococcus aureus, Staphylococcus epidermidis, Proteus mirabilis, Escherichia coli, Serratia marcescens, Moraxella catarrhalis, Enterobacter aerogenes, Enterobacter cloacae, Citrobacter freundii, Citrobacter koseri, Stenotrophomonas maltophilia, Bacteroides fragilis, Prevotella intermedia, Candida albicans, Aspergillus spp, Mucor spp, cytomegalovirus, herpes simplex virus type 1, influenza A and B viruses, respiratory syncytial virus, coronaviruses, rhinoviruses, parainfluenza viruses, and adenoviruses. In three positive samples, culture and PCR methods were negative for all bacterial, fungal, and viral agents (only PCR test performed for viral agents), except for one sample that had co-infection with Legionella pneumophila (Fig. 1).
The patient infected with Mimivirus-53 was a 25-year-old woman with renal disease, and her signs included fever, 10-kg weight loss, cough, night sweats, chest pain, and shortness of breath. The patient had been mechanically ventilated for more than eight days in the intensive care unit (ICU). Her clinical parameters showed an elevated white blood cell (WBC) count, erythrocyte sedimentation rate (ESR), and C-reactive protein (CRP) level. The level of vitamin D3 was below normal (Table 1). Computed tomography scan (CTS) indicated consolidation of the left lower lobe and bilateral basilar infiltrates. All pneumonia agents were negative for this patient. Out of three sputum samples, two samples were positive for Mimivirus, and the cycle threshold (Ct) value was 22 in the samples (Supplementary Fig. 3). Treatment with azithromycin was initiated for the patient, but the symptoms did not improve after 20 days, and she recovered slowly.
The patient with Mimivirus-186 was a diabetic 64-year-old man, presenting with fever, dyspnea, cough, and weight loss. He was diagnosed with pulmonary tuberculosis 5 years ago. The physician speculated the recurrence of Mycobacterium tuberculosis (M.tb) infection, but the results of smear, culture, and PCR tests for M.tb were negative. Also, all pneumonia agents were negative in the patient. He had an acute respiratory failure with the elevation of WBC count, CRP, and ESR (Table 1). CTS indicated peripheral subpleural opacity. After the evaluation of three BAL samples, all of them were found to be positive for Mimivirus. Also, the Ct values for BAL samples ranged from 18 to 20 (Supplementary Fig. 3).
The patient infected with Mimivirus-1291 was a 40-year-old man with human immunodeficiency virus (HIV), CD4 cell count below 300 cells/μL, and hepatitis B virus. Six months ago, he was admitted for treatment of Pneumocystis carinii pneumonia and oral candidiasis. During treatment, he had been mechanically ventilated for 12 days in the ICU. After 6 months of treatment, he complained of cough, fever, and night sweats with excessive sputum. Three sputum and two BAL samples were evaluated for pneumonia agents, but all important agents were negative. Two out of three sputum samples and two BAL samples were positive for Mimivirus infection. The Ct values were 19 and 21 for BAL and sputum samples, respectively (Supplementary Fig. 3). CTS indicated bilateral basilar infiltrates, suggesting viral pneumonia. Also, he showed increased WBC count, ESR, and CRP levels (Table 1).
The patient infected with Mimivirus-1922 was a 12-year-old boy with cystic fibrosis (CF). The CF genotype indicated a p.F508del mutation (ΔF508), and he was diagnosed with severe CF-related pulmonary disease. He was frequently admitted to the ICU and had been ventilated several times for a bacterial infection. Six months after discharge from the hospital, he showed signs of sputum production, weakness, night perspiration, cough, and fever. His biological parameters indicated elevated WBC count, ESR, and CRP levels (Table 1). CTS indicated multifocal consolidative changes and bilateral basilar infiltrates. The samples were negative for all pneumonia agents, except Legionella pneumophila. He was treated with azithromycin and levofloxacin, but his signs did not improve. All samples were evaluated for Mimivirus infection. Two BAL and two sputum samples with Ct values of 21 and 27 were positive for this virus, respectively, and the patient died unfortunately (Supplementary Fig. 3).
Discussion
To the best of our knowledge, the current study was the first to investigate the presence of Mimivirus in TB-suspected patients with pulmonary signs.
In nearly 20–50% of patients, the causative agent of pneumonia, as the leading cause of infection-related mortality worldwide, is unknown. Therefore, a major public health goal is to distinguish new agents, causing both community- and hospital-acquired pneumonia5. Moreover, the correlation between human diseases and viral infections needs to be evaluated, as the cause of many pneumonia cases is not yet determined10.
Water-borne pathogens colonize water sources in hospitals, and some of them may be correlated with amoeba, such as APMV10. According to several reports, the presence of Mimivirus-specific antibodies is more common in nosocomial pneumonia patients, particularly in ICUs, compared to the controls5,10. Also, patients with community-acquired pneumonia and serological evidence of APMV are usually re-hospitalized after discharge, perhaps owing to insufficient administration of drugs against viral infections10.
According to several studies, even if controversial, there is some evidence that Mimiviruses may be a causative agent of pneumonia, especially in patients, who are hospitalized in ICUs and are mechanically ventilated for a long time5,10,11. To confirm this hypothesis, the results of the present study indicated that three out of four patients were hospitalized in ICUs and mechanically ventilated. Also, several serological reports indicated the higher percentage of seroconversion to Mimiviruses in ventilator-associated pneumonia patients, compared to patients without ventilator-associated pneumonia11,12.
In the current study, Mimivirus was accompanied by Legionella pneumophila in one patient with CF. Overall, contamination with microbial agents of water supplies has been associated with both hospital- and community-acquired pneumonia outbreaks13. Bacteria, such as Burkholderia, Acinetobacter, Stenotrophomonas, Legionella, and Pseudomonas species, have been detected in hospital water supplies. Some of these species, including Legionella pneumophila, have been correlated with hospital- and community-acquired pneumonia, and also have been related to free-living amoebas in hospital and natural aquatic environments14,15,16. A previous study reported that patients with nosocomial pneumonia, who received care next to a contaminated water distribution system, had antibodies against amoeba pathogens17. Also, seroconversion to Acanthamoeba-surviving bacteria was detected in 40% of patients, hospitalized in ICUs, in association with ventilator-associated pneumonia, particularly in the absence of identified infectious agents. Mimivirus, as a member of the group of intra-amoebal microorganisms, could be considered a potential agent of pneumonia18. In the present study, one of the patients died, although he received anti-Legionella drugs. Co-infection with Mimivirus may be influential, as the other three patients, despite an underlying disease, recovered after a period.
Interestingly, our patients had underlying diseases, such as CF, HIV with a low CD4 count, diabetes, and renal disease. However, there is no evidence indicating the relationship between Mimivirus pulmonary infections and underlying diseases. The results of this study showed that individuals with an underlying disease were more likely to be infected with a pulmonary disease due to Mimivirus. However, Mimiviruses are potential pneumonia agents in both non-immunocompromised and immunocompromised patients19.
There are three lineages of Mimivirus (A, B, and C), according to the phylogenomic information (using conserved core genes)1. Two previous studies have reported that the lineage C of Mimivirus could cause a pulmonary disease8,20,21. In agreement with these results, all isolated Mimiviruses in this study were also from lineage C. It seems that lineage C of Mimivirus might be responsible for pulmonary infections.
In line with a study by La Scola5, besides real-time PCR assays, the suicide nested-PCR method was used in our study to avoid sample contamination while detecting this rare pathogen. Few studies have shown the presence of Mimivirus DNA in respiratory samples5,8,21. In the current study, the virus was detected in both lower and upper respiratory tracts by molecular techniques. Conversely, in several studies, PCR assays could not confirm that Mimivirus could be a human pathogenic agent.
In this regard, Dare et al.7 evaluated 496 respiratory specimens for Mimivirus by real-time PCR method, but no positive results were obtained for Mimivirus DNA. Another study evaluated Mimivirus DNA in 214 nasopharyngeal aspirate samples from children with bronchiolitis. This virus was not found in any of the respiratory samples22. One of the reasons for the discrepancy between the results could be the small sample size of these studies, compared to our study, besides the significant presence of nucleotide polymorphisms of giant virus genes; this hypothesis was confirmed by the current identification of many Mimiviruses23.
The limitation of this study was the lack of Acanthamoeba polyphaga strains for preparing antigens for a microimmunofluorescence study.
In conclusion, the results of the current study indicated that the prevalence of Mimivirus was 0.04% in TB-suspected patients. Our findings revealed that Mimivirus was correlated with pneumonia and that it was a potential agent of pneumonia in immunocompromised patients. However, further studies are necessary to confirm the role of Mimivirus pathogenicity in humans.
Data availability
All data that support all the experimental findings in this article is available in the Supplementary Data File provided. The Mimivirus partial genomes were submitted to GenBank under accession numbers, MN503292, MN503292, MN503294, and MN503295 for Mimivirus-53, Mimivirus-186, Mimivirus-1291, and Mimivirus-1922, respectively.
References
Colson, P., La Scola, B., Levasseur, A., Caetano-Anollés, G. & Raoult, D. Mimivirus: Leading the way in the discovery of giant viruses of amoebae. Nat. Rev. Microbiol. 15, 243 (2017).
La Scola, B., De Lamballerie, X., Claverie, J., Drancourt, M. & Raoult, D. Genus Mimivirus. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses, 275–276 (2005).
La Scola, B. et al. A giant virus in amoebae. Science 299, 2033–2033 (2003).
La Scola, B. et al. The virophage as a unique parasite of the giant mimivirus. Nature 455, 100 (2008).
La Scola, B., Marrie, T. J., Auffray, J.-P. & Raoult, D. Mimivirus in pneumonia patients. Emerg. Infect. Dis. 11, 449 (2005).
Khan, M., La Scola, B., Lepidi, H. & Raoult, D. Pneumonia in mice inoculated experimentally with Acanthamoeba polyphaga mimivirus. Microb. Pathog. 42, 56–61 (2007).
Dare, R. K., Chittaganpitch, M. & Erdman, D. D. Screening pneumonia patients for mimivirus. Emerg. Infect. Dis. 14, 465 (2008).
Saadi, H. et al. First isolation of Mimivirus in a patient with pneumonia. Clin. Infect. Dis. 57, e127–e134 (2013).
Assis, F. L. et al. Genome characterization of the first mimiviruses of lineage C isolated in Brazil. Front. Microbiol. 8, 2562 (2017).
Berger, P. et al. Ameba-associated microorganisms and diagnosis of nosocomial pneumonia. Emerg. Infect. Dis. 12, 248 (2006).
Raoult, D., Renesto, P. & Brouqui, P. Laboratory infection of a technician by mimivirus. Ann. Intern. Med. 144, 702–703 (2006).
Vincent, A. et al. Clinical significance of a positive serology for mimivirus in patients presenting a suspicion of ventilator-associated pneumonia. Crit. Care Med. 37, 111–118 (2009).
Anaissie, E. J., Penzak, S. R. & Dignani, M. C. The hospital water supply as a source of nosocomial infections: A plea for action. Arch. Intern. Med. 162, 1483–1492 (2002).
Barker, J. & Brown, M. Trojan horses of the microbial world: Protozoa and the survival of bacterial pathogens in the environment. Microbiology 140, 1253–1259 (1994).
Rowbotham, T. J. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33, 1179–1183 (1980).
Rutala, W. A. & Weber, D. J. Water as a reservoir of nosocomial pathogens. Infect. Control Hosp. Epidemiol. 18, 609–616 (1997).
La Scola, B., Mezi, L., Auffray, J.-P., Berland, Y. & Raoult, D. Patients in the intensive care unit are exposed to amoeba-associated pathogens. Infect. Control Hosp. Epidemiol. 23, 462–465 (2002).
La Scola, B. et al. Amoeba-resisting bacteria and ventilator-associated pneumonia. Emerg. Infect. Dis. 9, 815 (2003).
Vincent, A., La Scola, B. & Papazian, L. Advances in Mimivirus pathogenicity. Intervirology 53, 304–309 (2010).
Saadi, H. et al. Shan virus: A new mimivirus isolated from the stool of a Tunisian patient with pneumonia. Intervirology 56, 424–429 (2013).
Sakhaee Fatemeh, V. F., Golnaz, B., Davar, S. S. & Abolfazl, F. Pulmonary infection related to mimivirus in patient with primary ciliary dyskinesia. Emerg. Infect. Dis. 26, 1613 (2020).
Larcher, C., Jeller, V., Fischer, H. & Huemer, H. Prevalence of respiratory viruses, including newly identified viruses, in hospitalised children in Austria. Eur. J. Clin. Microbiol. Infect. Dis. 25, 681–686 (2006).
La Scola, B. et al. Tentative characterization of new environmental giant viruses by MALDI-TOF mass spectrometry. Intervirology 53, 344–353 (2010).
Acknowledgements
We would like to thank all of the patients who participated in the study.
Author information
Authors and Affiliations
Contributions
F.S., J.M.A., S.R., S.M., and F.A.: Performed the experiments and data acquisition; F.V., F.R.J., and S.D.S.: analyzed data and interpreted data; A.F.: designed and supervised clinical study, interpreted data, read and approved manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sakhaee, F., Mosayebi Amroabadi, J., Razi, S. et al. Detection of Mimivirus from respiratory samples in tuberculosis-suspected patients. Sci Rep 12, 8676 (2022). https://doi.org/10.1038/s41598-022-12757-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-12757-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.