Abstract
Risk stratification for hospitalized adults with COVID-19 is essential to inform decisions about individual patients and allocation of resources. So far, risk models for severe COVID outcomes have included age but have not been optimized to best serve the needs of either older or younger adults. Models also need to be updated to reflect improvements in COVID-19 treatments. This retrospective study analyzed data from 6906 hospitalized adults with COVID-19 from a community health system across five states in the western United States. Risk models were developed to predict mechanical ventilation illness or death across one to 56 days of hospitalization, using clinical data available within the first hour after either admission with COVID-19 or a first positive SARS-CoV-2 test. For the seven-day interval, models for age ≥ 18 and < 50 years reached AUROC 0.81 (95% CI 0.71–0.91) and models for age ≥ 50 years reached AUROC 0.82 (95% CI 0.77–0.86). Models revealed differences in the statistical significance and relative predictive value of risk factors between older and younger patients including age, BMI, vital signs, and laboratory results. In addition, for hospitalized patients, sex and chronic comorbidities had lower predictive value than vital signs and laboratory results.
Similar content being viewed by others
Introduction
The number of global confirmed cases with severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) infection has surpassed 257 million as of December 10, 2021, with over 5 million reported deaths1. Although the majority of patients infected by SARS-CoV-2 present with mild symptoms, studies reported that 20% get hospitalized and 5% of patients with Coronavirus disease 2019 (COVID-19) become critically ill2,3. From early on of the pandemic, both age and chronic comorbidities have been reported as a significant risk factor for poor outcomes4,5, and evidence supports increased risk with hypertension, diabetes, chronic obstructive pulmonary disease, chronic renal disease, and cardiovascular conditions4,6,7. Although young patients have a lower prevalence of comorbidities than aging patients, the relative risk of fatal outcome in young patients with hypertension, diabetes and cardiovascular diseases has been shown to be higher than in elderly patients8,9. In addition, some studies show the patient population tends to be younger with the emergence of delta as the variant of concern in the U.S. with regional proportions being greater than 99% as of November 202110. Assessing risk for severe COVID-19 in specific age groups is complicated by both the heterogeneity of clinical presentation and age-related differences in the prevalence of chronic multimorbidities. A deeper understanding of risk factors for COVID-19 severity among different age subpopulations is needed, as well as practical, explainable risk stratification for bedside clinical decision support, research stewardship, and advancing our biomedical understanding of SARS-CoV-2.
Several studies have described successful development of machine learning models to predict COVID-19 outcomes in hospitalized patients11,12,13,14,15,16,17,18,19,20. Further, explainable models can also inform care decisions by showing which factors lead a specific individual patient to be at risk for severe outcomes, and can also help show which variables are most important at the population level, suggesting areas for further research investigation21. However, existing studies have several limitations; (1) most are based on small sample sizes from academic centers, (2) higher incidence of severe outcomes in hospitalized cohorts than are typically observed with current treatments, (3) reliance on laboratory tests that are not routinely administered to all patients, (4) lack of investigation of differences in risk factors between younger and older hospitalized patients, and (5) marginal model performance for either of age groups13. To address these limitations, we develop high-performing age-stratified machine-learning models to predict the severity of COVID-19 progression from 6,906 patients in community hospitals across a large geographic area in the western United States, during five months after the delta variant had become predominant and new standards of care had lowered the severe outcome incidence rate. In addition, the model was developed to require only those laboratory results that are routinely administered for all COVID-19 patients.
Methods
Study design and setting
This retrospective study analyzed data gathered from Providence St. Joseph Health (PSJH), a community health system with 51 hospitals and 1085 clinics across five states in the western United States: Alaska, California, Montana, Oregon, and Washington. Inclusion criteria was age ≥ 18 years and confirmation of COVID-19 by a positive PCR-based SARS-CoV-2 test result. This study was performed in compliance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule and was approved by the Institutional Review Board (IRB) at PSJH with Study Number STUDY2020000196 with waiver of consent. We follow STROBE reporting guidelines (Supplemental Table S6).
Task definition
In this study, we hypothesized that age-stratified risk models for hospitalized patients with COVID-19 can accurately predict critical illness and mortality due to COVID-19 based on readily available patient data. Outcomes of patients were defined using the World Health Organization Ordinal Scale (WOS), proposed by the WHO R&D Blueprint group in their COVID-19 Therapeutic Trial Synopsis22. The WHO ordinal scale ranges from 0 (uninfected) to 8 (deceased) with gradations depending on hospitalization, supplemental oxygen, mechanical ventilation, and organ support (vasopressors, renal replacement therapy, and extracorporeal membrane oxygenation). See Supplemental Table S1. In this study, we categorized WHO ordinal scores of 3–5 as the mild cases of COVID-19 and WHO ordinal scores of 6–8 as the critical illness and death within hospitalized patients. The objective is to develop machine learning models to predict critical illness and death with COVID-19 in hospitalized patients using easily available variables, including aggregated laboratory biomarkers and vital signs within one hour of either admission to the hospital or the first positive inpatient SARS-CoV-2 test. These predictive models are developed on time horizons for one, seven, 14, 28, and 56 days from the confirmation of the infection and hospitalization to test the assumption that the baseline data up to one hour after hospital admission with a positive SARS-CoV-2 test can predict risk of critical illness on different time horizons. Additionally, we compare the performance of machine learning models within 7-days from the confirmation of the infection and hospitalization for (1) all-ages population, and (2) age-stratified subpopulations, to report the effect of age and compare the relative importance of risk factors between younger and aging adults.
Population
The start time point of study is defined as June 31, 2021, after the delta became the predominant SARS-CoV-2 variant in the Western United States. Studied population included hospitalized individuals who received a positive test for COVID-19 between June 31, 2021 and November 15, 2021. This was confirmed by reverse-transcriptase polymerase chain reaction (RT-PCR) for the SARS-CoV-2 ribonucleic acid (RNA). Patients were excluded if they were already receiving mechanical ventilation at the time of admission to the hospital.
Variables
The factors analyzed for prediction of COVID-19 outcomes were demographic characteristics, medical history, vital signs, and laboratory biomarkers (n = 64). We extracted the Charlson Comorbidity Index23 (CCI: measure of overall comorbidities) and individual chronic conditions that are known risk factors for poor COVID-19 outcomes (reported in the literature6) and conditions which are prevalent in aging patients (Table 1). Comorbidities that are usually chronic, such as hypertension, were included if they were active at the time of admission. Other comorbidities were included if they had been active any time within 2 years prior to admission, except for malignancy, which was included if active any time within the past 5 years. Note that active conditions mean health issues that affect the individual's current functioning and all health. We used ICD-10-CM (International Classification of Diseases, Tenth Revision, Clinical Modification) codes, which are shown together with SNOMED–CT© hierarchical parent codes (Supplemental Table S2). Laboratory results and vital signs were included (both inpatient and outpatient) if they were collected between 24 h before and one hour after either admission to the hospital or the first positive inpatient SARS-CoV-2 test (Table 1). Note that, we used aggregated temporal longitudinal vital signs in our model as described in Lee, et.al24. Additionally, the risk factor list included patients' need for supplemental oxygen mode, need for vasopressors, total number of comorbidities, and COVID-19 vaccination status.
Statistical analysis
Descriptive analyses are presented as frequencies and percentage for categorical variables, and as mean and standard deviation (std) for numerical variables. Fisher exact test was applied to compare distributions of categorical variables. The differences between distributions of numerical variables were calculated using Mann Whitney U-test. All statistical analyses were completed using PySpark version 2.4.5.
Risk model development
In data preprocessing for development of each risk model, we removed features with missing values greater than 20% (Supplemental Table S3). We used IterativeImputer from Scikit Learn version 0.24.0 for imputing missing data in numerical features25. Missing values for comorbidities were assumed to be absent from the patient’s medical history and imputed with a constant number of 0. Outliers were detected by calculating the modified z-score based on median absolute deviation with a threshold of 3.5 and then these outliers were imputed by the median.
To build machine learning models, we randomly split the dataset into 80% training data and 20% testing data and analyzed each patient using multiple algorithms including logistic regression (LR), random forest classification (RF), Adaptive Boosting (AdaBoost), and Gradient Boosting Decision Tree (GBDT). The parameters for each model were optimized using a tenfold cross-validation on the training set with the maximum scoring value for the area under receiver operating characteristic curve score (AUROC). We then balanced true and false positive rates by optimizing the probability threshold for each class. This optimal cut-off point is defined using the Youden index to maximize the summation of true positive rate and true negative rate.
To address collinearity between predictors, we compared the optimum performance of logistic regression using the least absolute shrinkage and selection operator (LASSO) feature selection method. For non-linear tree-based models all features were included. Performance of models was reported as the area under the receiver operating characteristic curve (AUROC), area under the precision-recall curve (AUPRC), true positive rate (TPR), true negative rate (TNR), predictive positive value (PPV), and negative predictive value (NPV). We reported the 95% confidence interval for performance metrics of the models using Wilcoxon statistics26, and binomial interval27 for the area under the ROC and precision-recall curves, respectively. All ML models were applied using Spark version 2.4.5, in the Python interface. We presented the interpretation of the model with the highest relative performance, gradient boosting, using the Shapley additive explanations (SHAP) algorithm, which uses cooperative game theory to calculate the marginal contribution of each feature, and examines the feature influence on model prediction28. Predictive models were reported following TRIPOD guidelines29.
Results
Baseline characteristics
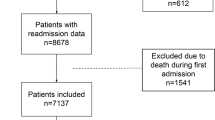
In the Providence St Joseph cohort (described in “Methods” above), 6,906 patients with positive tests for SARS-CoV-2 were analyzed (Supplemental Fig. S1). The severe outcome incidence rate of 10.88%. Percent female was 44.25 and mean age was 59.90 years (SD ± 17.83 years), with a range 18 to 90 + years old. The distribution of relative frequency of hospitalizations by age is shown in Supplemental Fig. S2. We divided the patients into two age subgroups: younger (age ≥ 18 and < 50 years with 1,963 patients) and older (≥ 50 years with 4,943 patients). The reported variables for prognosis of COVID-19 critical illness are presented in Table 2, Supplemental Table S3, and Supplemental Table S4. For patients with age ≥ 18 and < 50 years, the variables that had statistically significant correlation with critical illness and death in patients with COVID-19 were BMI, age, heart failure, and cardiomyopathy. For patients with age ≥ 50 years, the statistically significant variables were BMI, age, sex, dementia, and use of vasopressors within one hour of either admission to the hospital with COVID-19 or a first positive inpatient SARS-CoV-2 test. Vital signs values were aggregated from 24 h before to one hour after and included (mean and standard deviation) for heart rate (HR), systolic blood pressure (SBP), diastolic blood pressure DBP, respiratory rate (RR), blood oxygen saturation (SpO2), and body temperature.
Risk model analysis
In this paper, we trained five ML models including LR, RF, GBDT, and AdaBoost for the all-age population (n = 6,906), and two different age subpopulations (patients with age ≥ 18 and < 50 years with n = 1,963 and patients with age ≥ 50 years with n = 4,943) using the aggregated values of predictors. Class distribution for outcomes show that patients with critical illness and death accounted for 7.79% of the younger cohort with age ≥ 18 and < 50 years and 12.04% of the older cohort with age ≥ 50 years. This class imbalance was addressed by undersampling patients with mild severity from the training set. Results were reported on the complete test dataset, representing actual population distribution. Supplemental Table S5 represents the performance results for three sets of developed models for younger, older patients and all-age groups. These performance results were reported after adjusting the probability threshold to optimize models for clinical and research applications. Among four models for the younger population, GBDT had the highest true positive rate of 74.98%, true negative rate of 74.04%, and AUROC value of 0.78. For the older population, GBDT had a maximum true positive rate of 72.72%, true negative rate of 72.91% and AUROC of 0.81. Figure 1 represents the comparison between the AUROC values for four ML models based on the patient's age. Relative feature importance for the younger, older and generalized GBDT models was determined by Shapley additive explanations (SHAP), as shown in Fig. 2, and Supplemental Fig. S5, respectively. SHAP values were also used to assess the contribution of age on each model outcome (Supplemental Fig. S5). In addition, Supplemental Fig. S6 presents the model with individual comorbidities as risk factors for age-stratified models. We used the distribution of importance for each variable to assess its contribution to model outcome. In the younger population, some variables for comorbidities added no predictive value, which resulted in them being automatically removed from the SHAP plot.
Gradient Boosting Decision Tree feature importance for age-stratified models of severe COVID-19 outcomes in hospitalized patients. (A) Feature importance and the influence of higher and lower values of the risk factors on the patient with age ≥ 18 and < 50 years outcome, (B) Feature importance and the influence of higher and lower values of the risk factors on the patient with age ≥ 50 years outcome. Note that the left side of this graph represents reduced risk of critical illness or death, and the right side of the graph represents the increased risk of critical illness and death outcome. Nominal classes are binary [0, 1]. For sex, female is 0 (blue) and for race, White is 0 (blue).
Additionally, we used the GBDT to validate and assess the performance of the model for different time horizons. For the all-age group, gradient boosting showed an AUROC value of 0.83, 0.80, 0.79 and 0.79 for respectively, 1-, 14-, 28- and 56-day intervals after the confirmation of infection. Furthermore, we predicted the mortality of patients (WHO ordinal score of 8) using the GBDT model and the full set of aggregated risk factors. Note that to predict the mortality of patients with COVID-19, we also included the patients who were already receiving mechanical ventilation and additional organ support (WHO ordinal score of 6 and 7), see Supplemental Fig. S2. Therefore, the number of all age group patients for predicting mortality increased to 7,063. The results show the AUROC value of 0.82 for the general population and 0.79 and 0.75 for the younger and aging population, respectively.
Discussion
In this study, we developed risk models to predict the outcomes of hospitalized adult patients with COVID-19, in the context of current COVID-19 standard of care and delta variant predominance. We used clinical data from within one hour of either admission to the hospital with COVID-19 or the first positive inpatient SARS-CoV-2 test result. Explainability analysis on the machine learning models showed that risk factors are different for older patients compared to younger patients. This is the first study that investigates age-stratified modeling for COVID-19 severity for hospitalized adults for early prediction across multiple time horizons. Data from 6,906 patients across five states was used to develop predictive models for COVID-19 critical illness and death in younger and older hospitalized adults within one, seven, 14, 28 and 56 days of positive infection test and hospitalization. The key findings are: 1) risk models perform well using readily available clinical data, 2) vital signs and laboratory results at the time of admission are more important for prediction than the presence of comorbidities, 3) age-stratified models show that the relative importance of risk factor differs between younger and older adults.
Since the beginning of the pandemic, standard of COVID-19 care has improved and delta has become the predominant variant. Further, risk models from earlier in the pandemic relied on labs that are not routinely used in many patients. This was reflected by the high rate of missing values for tests required for in early risk scores, including INR, D-dimer, ferritin and procalcitonin (PCT). The models developed here are both performant and pragmatic.
Our statistical analysis revealed new insights on how variables that correlate significantly with critical illness and death in COVID-19 differ between younger and older age groups. For example, most comorbidities such as malignancy, cardiomyopathy and COPD have higher odds ratios for severe outcomes in younger patients than in older patients. Conversely, lower BUN/creatinine ratio and lower potassium are only statistically associated with critical illness and death in older patients.
We chose GBDT, a sequential ensemble approach30, as the model with the best relative performance to define the most predictive variables for COVID-19 outcomes. Non-linear models showed higher performance than linear models, suggesting better representation of complex interactions across multiple mechanisms of disease. Stratifying patients by age group revealed that, in general, vital signs and laboratory tests have a higher relative importance than comorbidities. Because age is such a significant risk factor, it can mask other important predictors. By removing the confounding effects of age, these models highlight new insights into risk factors for IMV and death.
Additionally, we investigated the effect of age on predictive models for younger and older COVID-19 patients. For patients with age ≥ 18 and < 50 years (Supplemental Fig. S5C), age has a relatively high and more consistent predictive effect on the performance of the model. Within patients younger than 50 years old, higher age had a negative effect on outcome. However, in patients with age ≥ 50 years (Supplemental Fig. S5D), age has less effect on the model performance. Patient stratification removed some of the confounding effect of age in this group, better revealing the contribution of laboratory results, vital signs and comorbidities as predictors.
For the younger population, the patient's initial oxygen mode and aggregated vital signs demonstrate the highest predictive value for outcome severity. Other predictive factors include higher AST, higher creatinine, and lower calcium levels, higher age, and higher BMI. Laboratory results have higher importance for older patients than they do for younger patients. Features such as higher BUN, higher AST, lower HCO3, lower calcium, and some aggregated vital signs (respiratory rate, blood pressure and SpO2) are among the most predictive. Sex is not a strong predictive factor, despite it having an odds ratio of ~ 1.25 in both the older and younger population. BMI is another feature that supports the importance of analyzing age subgroups separately. It is statistically correlated to the severity of COVID-19 and is an important predictor for the younger population but shows no significant correlation in the older population (Supplemental Fig. S4). This could be explained by higher BMI in younger hospitalized patients compared to the older hospitalized patients with COVID-1931. Future investigation is needed to determine risks with being underweight or overweight, potentially with BMI-stratified models. Neither race nor ethnicity had strong feature importance for prediction in the younger and older population. This shows that although chronic comorbidities (Charlson Comorbidity Index or binary diagnostic labels), sex, race, ethnicity may have high odds ratios in a univariate analysis, these factors are much less important in the acute setting for predicting critical illness. Once hospitalized, biomedical observations are more predictive. Chronic conditions are still important for predicting the severity of COVID-19 outcomes, but medical and clinical biomarkers have a higher predictive value. The importance of comorbidities and CCI has also been investigated by comparing a predictive model which includes only demographics and CCI. The comparison of models performance is presented in Supplementary Fig. S6 and SHAPs are presented in Supplementary Fig. S7 and Supplementary Fig. S8.
SHAP values also indicate the direction of variables’ impact on outcomes. For example, higher serum creatinine levels, lower platelet counts, lower lymphocyte counts, and higher neutrophil count are all predictive of critical illness and death28 Lower calcium is associated with more severe COVID-19, as noted in previous studies32, and this analysis shows it has higher predictive value in older patients.
Hence, age stratification shows that risk factors for severe COVID-19 differ by age, in ways that cannot be determined in all-age models. This affirms the importance of analyzing each different age group separately, particularly for the older population who have the greater overall risk for poor outcomes.
Also, as expected, vaccination reduced the risk of severe outcomes in the older population. Vaccination status had relatively low importance, which may reflect the low number of hospitalized patients who had received vaccination during the observation window; only 8.10% of the younger hospitalized patients and 25.48% of older hospitalized patients had received at least one dose of a vaccine (Supplemental Fig. S3).
Early risk stratification in patients with COVID-19 is essential to inform decisions about what level of care a patient is likely to need. One of the main challenges of COVID-19 is the heterogeneity of presentation; therefore factors related to poor outcomes are not always evident at admission15. In this study, ML models using readily available variables (demographics, vital signs, common laboratory test and medical history) demonstrated strong performance for predicting the severity of COVID-19. Importantly, the population in this study included patients from 51 hospitals and 1081 clinics across five states, using data based on the current standard of care for COVID-19 and the delta variant. Five limitations of this retrospective study are: 1) reliance on EHR structured data which can miss medical conditions that not diagnosed, not recorded, or noted only in free text, 2) use of hospital reported race and ethnicity of patients33 as opposed to direct per-patient measures of potential confounders (genetic information, disparities in healthcare, and individual lifetime history of beneficial and harmful exposures, 4) use of data from within a single healthcare system. Concerns regarding generalizability of this study are partially mitigated by the size and diversity of PSJH, which serves both urban and rural communities from California to Alaska. Future investigations will benefit from finer granularity of subdivisions by age, BMI, and more detailed variables on conditions and drugs that affect individual immune response.
Conclusion
We developed two age-stratified risk models for critical illness in hospitalized patients with COVID-19 and tested them on data from patients during times of improved standard of care treatment and delta variant predominance. For hospitalized adults, baseline data that is readily available within one hour after hospital admission or a first positive inpatient SARS-CoV-2 test can predict critical illness within one day, and up to 56 days later. The models for age ≥ 18 and < 50 years and the model for age ≥ 50 years were both more performant than all-age models. These age-stratified models also revealed differences in the statistical significance and relative predictive value of risk factors between older and younger patients, including age, BMI, vital signs, and laboratory results. In addition, sex and chronic comorbidities had lower predictive value than vital signs and laboratory results. The results of this age-stratified modeling approach provide advanced understanding of current risk factors for severe COVID-19 outcomes and can help inform care decisions and prioritize next steps for research.
Data availability
All clinical logic has been shared within the paper and supplemental materials. Results have been aggregated and reported within this paper to the extent possible while maintaining privacy from personal health information as required by law. Data are archived within Providence St Joseph Health systems in a HIPAA-secure audited compute environment. For information, contact the Vice President of Information Management at Providence St Joseph Health.
References
WHO Coronavirus Disease (COVID-19) Dashboard at https://covid19.who.int. Accessed 10 Dec 2021.
Bohn, M. K. et al. Pathophysiology of COVID-19: Mechanisms underlying disease severity and progression. Physiology 35, 288–301 (2020).
Joost Wiersinga, W., Rhodes, A., Cheng, A. C., Peacock, S. J. & Prescott, H. C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): A review. JAMA 324, 782–793 (2020).
Zhou, F. et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 395, 1054–1062 (2020).
Kim, L. et al. Risk factors for intensive care unit admission and in-hospital mortality among hospitalized adults identified through the us coronavirus disease 2019 (COVID-19)-associated hospitalization surveillance network (COVID-NET). Clin. Infect. Dis. 72, e206–e214 (2021).
Gao, Y.-D. et al. Risk factors for severe and critically ill COVID-19 patients: A review. Allergy 76, 428–455 (2021).
Farshbafnadi, M., Kamali Zonouzi, S., Sabahi, M., Dolatshahi, M. & Aarabi, M. H. Aging & COVID-19 susceptibility, disease severity, and clinical outcomes: The role of entangled risk factors. Exp. Gerontol. 154, 111507 (2021).
Bae, S., Kim, S. R., Kim, M.-N., Shim, W. J. & Park, S.-M. Impact of cardiovascular disease and risk factors on fatal outcomes in patients with COVID-19 according to age: a systematic review and meta-analysis. Heart 107, 373–380 (2021).
Goodman, K. E., Magder, L. S., Baghdadi, J. D., Pineles, L., Levine, A. R., Perencevich, E. N. & Harris, A. D. Impact of Sex and Metabolic Comorbidities on Coronavirus Disease 2019 (COVID-19) Mortality Risk Across Age Groups: 66 646 Inpatients Across 613 U.S. Hospitals. Clin. Infect. Dis. 73, e4113–e4123 (2021).
Burki, T. K. Lifting of COVID-19 restrictions in the UK and the Delta variant. Lancet Respir Med 9, e85 (2021).
Marcos, M., Belhassen-García, M., Sánchez-Puente, A., Sampedro-Gomez, J., Azibeiro, R., Dorado-Díaz, P.-I., Marcano-Millán, E., García-Vidal, C., Moreiro-Barroso, M.-T., Cubino-Bóveda, N., Pérez-García, M.-L., Rodríguez-Alonso, B., Encinas-Sánchez, D., Peña-Balbuena, S., Sobejano-Fuertes, E., Inés, S., Carbonell, C., López-Parra, M., Andrade-Meira, F., López-Bernús, A., Lorenzo, C., Carpio, A., Polo-San-Ricardo, D., Sánchez-Hernández, M.-V., Borrás, R., Sagredo-Meneses, V., Sanchez, P.-L., Soriano, A. & Martín-Oterino, J.-Á. Development of a severity of disease score and classification model by machine learning for hospitalized COVID-19 patients. PLoS One 16, e0240200 (2021).
Lombardi, Y. et al. External validation of prognostic scores for COVID-19: a multicenter cohort study of patients hospitalized in Greater Paris University Hospitals. Intensive Care Med. 47(12), 1426–1439 (2021).
King, J. T. Jr. et al. Accuracy of the Veterans Health Administration COVID-19 (VACO) Index for predicting short-term mortality among 1307 US academic medical centre inpatients and 427 224 US Medicare patients. J. Epidemiol. Commun. Health 76(3), 254–260 (2021).
Rinderknecht, M. D. & Klopfenstein, Y. Predicting critical state after COVID-19 diagnosis: Model development using a large US electronic health record dataset. NPJ Digit Med 4, 113 (2021).
Yadaw, A. S. et al. Clinical features of COVID-19 mortality: development and validation of a clinical prediction model. Lancet Digit Health 2, e516–e525 (2020).
Nicholson, C. J., Wooster, L., Sigurslid, H. H., Li, R. H., Jiang, W., Tian, W., Lino Cardenas, C. L. & Malhotra, R. Estimating risk of mechanical ventilation and in-hospital mortality among adult COVID-19 patients admitted to Mass General Brigham: The VICE and DICE scores. EClinicalMedicine 33, 100765 (2021).
Knight, S. R., Ho, A., Pius, R., Buchan, I., Carson, G., Drake, T. M., Dunning, J., Fairfield, C. J., Gamble, C., Green, C. A., Gupta, R., Halpin, S., Hardwick, H. E., Holden, K. A., Horby, P. W., Jackson, C., Mclean, K. A., Merson, L., Nguyen-Van-Tam, J. S., Norman, L., Noursadeghi, M., Olliaro, P. L., Pritchard, M. G., Russell, C. D., Shaw, C. A., Sheikh, A., Solomon, T., Sudlow, C., Swann, O. V., Turtle, L. C. W., Openshaw, P. J. M., Kenneth Baillie, J., Semple, M. G., Docherty, A. B. & Harrison, E. M. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ 370, (2020).
Chen, Z. et al. A risk score based on baseline risk factors for predicting mortality in COVID-19 patients. Curr. Med. Res. Opin. 37, 917–927 (2021).
Garcia-Gordillo, J. A., Camiro-Zúñiga, A., Aguilar-Soto, M., Cuenca, D., Cadena-Fernández, A., Khouri, L. S., Rayek, J. N., Mercado, M. & ARMII Study Group. COVID-IRS: A novel predictive score for risk of invasive mechanical ventilation in patients with COVID-19. PLoS One 16, e0248357 (2021).
Gong, J. et al. A tool for early prediction of severe coronavirus disease 2019 (COVID-19): A multicenter study using the risk Nomogram in Wuhan and Guangdong China. Clin. Infect. Dis. 71, 833–840 (2020).
Ahmad, M. A., Eckert, C. & Teredesai, A. Interpretable Machine Learning in Healthcare. in Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics 559–560 (Association for Computing Machinery, 2018).
Coronavirus, N. WHO R&D Blueprint. https://www.who.int/blueprint/priority-diseases/key-action/COVID-19_Treatment_Trial_Design_Master_Protocol_synopsis_Final_18022020.pdf. Accessed 10 January 2021.
Charlson, M. E., Pompei, P., Ales, K. L. & MacKenzie, C. R. A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J. Chronic Dis. 40, 373–383 (1987).
Lee, J. Y., Molani, S., Fang, C., Jade, K., Kornilov, S. A., Mico, L. T., O’mahony, S. & Hadlock, J. J. Ambulatory risk models for the long-term prevention of sepsis: Retrospective study. JMIR Med Inform 9, e29986 (2021).
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P., Weiss, R., Dubourg, V. & Others. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Hanley, J. A. & McNeil, B. J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 (1982).
Boyd, K., Eng, K. H. & Page, C. D. Area under the precision-recall curve: Point estimates and confidence intervals. in Machine Learning and Knowledge Discovery in Databases 451–466 (Springer Berlin Heidelberg, 2013).
Rodríguez-Pérez, R. & Bajorath, J. Interpretation of compound activity predictions from complex machine learning models using local approximations and shapley values. J. Med. Chem. 63, 8761–8777 (2020).
Collins, G. S., Reitsma, J. B., Altman, D. G. & Moons, K. G. M. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): The TRIPOD Statement. Ann. Intern. Med. 162, 55–63 (2015).
Friedman, J. H. Stochastic gradient boosting. Comput. Stat. Data Anal. 38, 367–378 (2002).
Bhasin, A. et al. Is BMI higher in younger patients with COVID-19? association between BMI and COVID-19 hospitalization by age. Obesity 28, 1811–1814 (2020).
di Filippo, L., Doga, M., Frara, S. & Giustina, A. Hypocalcemia in COVID-19: Prevalence, clinical significance and therapeutic implications. Rev. Endocr. Metab. Disord (2021).
Zingmond, D. S. et al. Improving hospital reporting of patient race and ethnicity-approaches to data auditing. Health Serv. Res. 50(Suppl 1), 1372–1389 (2015).
Acknowledgements
We are grateful to Providence St. Joseph Health for sharing their data engineering expertise and computational resources. We would also like to acknowledge SNOMED International for developing and maintaining SNOMED-CT.
Funding
This work was funded by NIH NIA Grant 2U01AG046139-06 (to NDP, NET). JJH and VRD have been funded in part with Federal funds from the Department of Health and Human Services, Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority, under Contract No. HHSO100201600031C, administered by Merck, Inc., on work unrelated to this study. NET is also funded by NIH NIA R01 AG061796. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
S.M., P.V.H., J.D.G., and J.J.H. conceptualized the study. S.M., R.T.R., P.V.H., V.R.D. and A.M.B. were involved in the EHR data extraction, data cleaning, and codification. S.M. performed data analysis including statistical analysis, machine learning and data interpretation. J.J.H. supervised implementation. Administrative and material support was provided by J.J.H. and N.D.P. S.M. and P.V.H. prepared the manuscript with critical revision of the manuscript for important intellectual content provided by J.D.G., N.R., and J.J.H. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
JJH and SM have received grant funding from Pfizer for research unrelated to this work. JDG declared contracted research with Gilead, Lilly, and Regeneron, and fees from Gilead and Lilly for speaking and advisory board. JJH declared grant funding from Pfizer for COVID-19 research unrelated to this study. None of the other authors declare a competing interest with this study.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Molani, S., Hernandez, P.V., Roper, R.T. et al. Risk factors for severe COVID-19 differ by age for hospitalized adults. Sci Rep 12, 6568 (2022). https://doi.org/10.1038/s41598-022-10344-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-10344-3
This article is cited by
-
The transition from genomics to phenomics in personalized population health
Nature Reviews Genetics (2024)
-
Association between any underlying health condition and COVID-19-associated hospitalization by age group, Washington State, 2020–2021: a retrospective cohort study
BMC Infectious Diseases (2023)
-
Demographics, clinical characteristics, and outcomes in hospitalized patients during six waves of COVID‑19 in Northern Iran: a large cohort study
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.