Abstract
Lineage tracing in mice indicates that LGR5 is an adult stem cell marker in multiple organs, such as the intestine, stomach, hair follicles, ovary, and mammary glands. Despite many studies exploring the presence of LGR5 cells in human tissues, little is known about its expression profile in either human mammary tissue or pathological lesions. In this study we aim to investigate LGR5 expression in normal, benign, and malignant lesions of the human breast using RNA in situ hybridization. LGR5 expression has not been observed in normal lactiferous ducts and terminal duct lobular units, whereas LGR5-positive cells have been specifically observed in the basal myoepithelium of ducts in the regenerative tissues, ductal carcinoma in situ, and in ducts surrounded by invasive cancer cells. These findings suggest LGR5 marks facultative stem cells that are involved in post injury regeneration instead of homeostatic stem cells. LGR5 positivity was found in 3% (9 of 278 cases) of invasive breast cancers (BC), and it showed positive associations with higher histologic grades (P = 0.001) and T stages (P < 0.001), while having negative correlations with estrogen receptor (P < 0.001) and progesterone receptor (P < 0.001) expression. Remarkably, all LGR5-positive BC, except one, belong to triple-negative BC (TNBC), representing 24% (9 of 38 cases) of all of them. LGR5 histoscores have no correlations with EGFR, CK5/6, Ki-67, or P53 expression. Additionally, no β-catenin nuclear localization was observed in LGR5-positive BC, indicating that canonical Wnt pathway activation is less likely involved in LGR5 expression in BC. Our results demonstrate that LGR5 expression is induced in regenerative conditions in the myoepithelium of human mammary ducts and that its expression is only observed in TNBC subtype among all invasive BC. Further studies regarding the functional and prognostic impact of LGR5 in TNBC are warranted.
Similar content being viewed by others
Introduction
Leucine-rich repeat containing G-protein-coupled receptor 5 (LGR5) encodes a seven-transmembrane receptor belonging to the G-protein-coupled receptor rhodopsin family. LGR5 and its close homologs, LGR4 and LGR6, are potent enhancers of canonical Wnt/β-catenin signaling by binding to secreted R-spondin growth factors1. In the absence of R-spondins, the E3-ubiquitin ligases Rnf43/Znrf3 degrades the Frizzled receptor, leading to downregulation of Wnt signaling2. As Rnf43/Znrf3 are themselves transcriptional Wnt/β-catenin signaling targets, they serve as components of a negative Wnt feedback loop2. LGR5 has been identified as a homeostatic stem cell exquisite marker in various tissues, including the intestines, stomach, hair follicles, ovaries, and mammary glands3,4,5,6,7. Subsequently, LGR5 + cells have also been demonstrated to be facultative stem cells responsible for postinjury regeneration in the liver, pancreas, and stomach8,9,10. Homeostatic LGR5 + stem cells contribute to various cancers such as colorectal cancers, gastric cancers, and squamous cell skin carcinomas when oncogenic mutations occur11,12,13.
Cancer stem cells are widely believed to be responsible for cancer initiation and progression. They are a small tumor population with stem cell properties. A growing number of studies demonstrate that CSCs are remarkably heterogeneous and plastic. Therefore, they can convert from differentiated cells under permissive conditions14. In colorectal cancers, LGR5 + cells have been demonstrated to act as cancer stem cells fueling tumor growth and metastasis15,16. In addition, Yang et al. suggested that LGR5 plays a key role in maintaining breast cancer (BC) stem‐like cells through Wnt/β‐catenin signaling17. There also exist several studies that have examined the prognostic significance of LGR5 in BC, and mostly they show an immunohistochemical staining to detect LGR5+ cells in cancer tissues17,18,19. However, it is well known that there are no reliable antibodies for marking LGR5 + cells with formalin-fixed paraffin-embedded (FFPE) tissues. Recently RNA in situ hybridization (ISH) techniques have been used to visualize LGR5 + cells in human tissues, and this has been proven to be successful in many types of cancers. For BC, Ogasawara et al. have demonstrated specific LGR5 mRNA expressions using an RNAscope in 43 tripe negative BC20. In this study, we aim to thoroughly investigate LGR5 expression in a large number of pathologic breast lesions, including not only invasive cancers but also a variety of benign lesions.
Material and methods
Subjects
We obtained BC tissues from 293 patients (278 invasive carcinoma and 15 DCIS cases) who had undergone surgical resection at Jeju National University Hospital between 2012 and 2019. We gathered clinical pathological information, including age, gender, size, tumor grade, presence of lymphovascular invasion, lymph node metastasis, American Joint Committee on Cancer/International Union against Cancer (AJCC/UICC) cancer staging (7th edition), and positivity for ER, PR, CK5/6, EGFR and HER2 from the patients’ medical records. BCs were subclassified according to ER, PR, and HER2 expression, luminal A, luminal B, HER2, and TNBCs. We also collected normal tissues and benign mammary lesions, including normal lobules (n = 5), lactiferous ducts (n = 5), fibroadenomas (n = 7), phyllodes tumors (n = 2), intraductal papillomas (n = 3), adenoses (n = 5), and inflammatory (n = 7) or post-biopsy or excision tissues (n = 3). This study was approved by the ethics committee of the Institutional Review Board of Jeju National University Hospital (IRB No.: 2019-04-006, “Expression analysis of LGR5 in breast cancer”) and was conducted in accordance with the Declaration of Helsinki. Informed consent from the patients was waived with IRB approval.
Tissue microarray construction
In total, 16 tissue microarrays (TMAs) were constructed from archival FFPE tissue blocks, including 293 primary BC tissues and 37 benign lesions. In brief, through histologic examination, a representative tumor portion was carefully selected from hematoxylin- and eosin-stained slides. Each tumor area comprised more than 70% of the cell population. The 4-mm diameter core tissues were obtained from individual BC paraffin blocks or benign lesions and arranged in a new recipient paraffin block (tissue array block) using a trephine apparatus (SuperBioChips Laboratories, Seoul, Korea).
Immunohistochemistry and interpretation
Immunohistochemistry (IHC) was done with the Ventana Benchmark Ultra platform (Ventana Medical Systems Inc., Tucson, AZ, USA); estrogen receptor (ER) (clone, SP1; Cat. No., 790-4324), progesterone receptor (PR) (clone, 1E2; Cat. No., 790-2223), HER2 (clone, SP3; Cat. No., 790-4493), CK5/6 (clone, D5&16B4; Cat. No., 790-4554), EGFR (clone, 3C6; Cat. No., 790-2988), P53 (clone, D0-7; Cat. No., 800-2912), P63 (clone, 4A4; Cat. No., 790-4509) and Ki-67 (clone, 30-9, Cat. No., 790-4286). HER2 expression was scored according to the 2007 ASCO/CAP guidelines: 0, no staining; 1 +, weak and incomplete membranous staining in ≥ 10% of the tumor cells; 2 +, weak-to-moderate complete membranous staining in ≥ 10% of the tumor cells; and 3 + , strong, complete membranous staining in ≥ 30% of the tumor cells21. HER2 was defined as positive when the IHC score is 3 or fluorescent in situ hybridization (FISH) is positive for the cases with IHC score 2. ER and PR were scored with the Allred system (range: 0–8); defined as being positive when it is more than 3. The intensity and percentage of EGFR and CK5/6 tumor cell expressions were measured by multiplying the intensity score (0 = negative; 1 = weak; 2 = moderate; 3 = strong) and percentage of positive cells (range = 0–100), ranging from 0 to 300. P53 and Ki-67 staining was recorded as the percentage of nuclear stained tumor cells. IHC for β-catenin was performed using a BOND-MAX automated immunostainer and a Bond Polymer Refine Detection kit (Leica Microsystems, Wetzlar, Germany) (clone, 17C2; Novocastra Laboratories, Newcastle, UK), and nuclear was considered as positive when more than 10% of tumor nuclei were stained.
LGR5 RNA in situ hybridization
We performed LGR5 mRNA detection using an RNAscope kit (Advanced Cell Diagnostics, Hayward, CA, USA) with unstained tissue slides according to the manufacturer's instructions. Tissue sections were pretreated with protease application and heating prior to hybridization with an LGR5-specific probe. The detailed procedure is described in an earlier publication17. Brown punctate dots present in the nucleus and/or cytoplasm indicated positive staining. LGR5 expression was quantified according to the five-grade scoring system recommended by the manufacturer (grade 0: no staining, 1: grade 1–3 dots/cell, grade 2: 4–10 dots/cell, grade 3: > 10 dots/cell, grade 4: > 15 dots/cell with > 10% of dots in clusters). The grade and percentage of tumor cells expressing LGR5 were measured, and histoscores (H-scores) were calculated by multiplying the grade (range = 1–4) and percentage of LGR5-positive tumor cells (range = 0–100), ranging from 0 to 400. For statistical analyses, the case was defined as being positive if H-scores are more than 10. For dual ISH for LGR5 and IHC for P63, IHC was conducted after completion of the in situ hybridization protocol.
Statistics
The SPSS (Statistical Package for the Social Sciences) statistical software version 18.0 (SPSS, Chicago, IL, USA) and Prism version 9.0.1 (GraphPad Software, San Diego, CA, USA) were used for analysis. We compared LGR5 H-scores between subtypes of invasive BC by using Tukey’s Multiple Comparison Test. We analyzed the LGR5 positivity clinical correlation study with the Pearson χ2 test. The correlations between the LGR5 H-scores and several molecular marker expressions were evaluated by the Spearman correlation test. Differences were considered significant when P < 0.05.
Results
LGR5 expression in normal breast lobules, benign lesions, and ductal carcinoma in situ
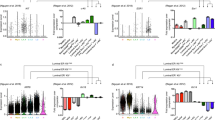
LGR5+ cells have consistently been observed in the basal myoepithelial cells of murine mammary ducts near the nipple7,22,23,24. To see whether LGR5-expressing cells exist in proximal human breast ducts, we collected five cases of lactiferous ducts. However, LGR5 expression was not observed in either the luminal or the basal cells (Fig. 1A). We investigated LGR5 expression in normal terminal duct lobular units (TDLU) and various benign lesions, including adenosis, intraductal papillomas, fibroadenomas, and phyllodes tumors. None of them showed LGR5 expression. Next, we examined preinvasive BC (15 DCIS cases) and LGR5 expression was focally observed in 9 cases (60%). Notably, LGR5 expression was completely restricted to the basal cells surrounding the DCIS, whereas no carcinoma cells expressed LGR5 (Fig. 1B and Supplementary Fig. S1). The overall positivity rates are summarized in Fig. 1C. These findings suggest that normally there are no LGR5-positive cells in adult human mammary ducts and lobules, but basal cells expressing LGR5 emerge in DCIS.
LGR5 Expression in Benign Lesions and Ductal Carcinoma In Situ (DCIS). (A) No LGR5 expression was observed in lactiferous ducts, terminal duct lobular unit, and adenosis. (B) Representative images of LGR5 expression in DCIS. Red arrows indicate LGR5-positive myoepithelial cells that express P63. (C) A table showing the percentages of LGR5 positivity in benign lesions and DCIS. H&E hematoxylin and eosin; No number; TDLU terminal duct lobular unit; IDP intraductal papilloma, FA fibroadenoma; PT phyllodes tumor. Scale bars: 50 μm.
LGR5 induction in myoepithelial cells of the regenerative mammary ducts
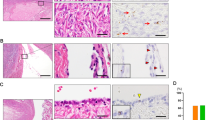
LGR5 cells have been identified as reserve stem cells in several adult murine organs, but only upon tissue injury for recovery. As LGR5 cells are not present in normal and benign mammary lesions, we explored whether they could be induced under regenerative conditions. Among the 10 inflammatory or healing lesions examined, we found two cases where LGR5 cells emerged in regenerative ducts. The first case was an excisional specimen of adenosis containing a scar area induced by the needle biopsy. We observed LGR5-positive cells in the linear ductal structures and β-catenin staining demonstrated that they were epithelial cells but not stromal cells (Fig. 2A). The other was a resected specimen where LGR5-positive cells were observed in the inflamed ducts around the excision site (Fig. 2B). To identify what type of cells express LGR5, we performed dual stain for LGR5 and P63 on the second case, and we confirmed that LGR5 cells are P63-positive myoepithelial cells (Fig. 2B). These findings suggest that in human mammary tissues, LGR5 cells can emerge under certain conditions such as regeneration following tissue injury.
Induced LGR5 Expression in Regenerative Tissues. (A) RNA in situ hybridization showed the LGR5-positive cells in the needle biopsy-induced scar area in adenosis (indicated by red arrows) and β-catenin staining confirmed that they are epithelial cells (indicated by black arrows). (B) Regenerative areas after excisional biopsy shows a group of LGR5-positive cells mixed with inflammatory cells. Dual staining for P63 (brown nuclear stain) and LGR5 (brown dots in the cytoplasm) demonstrated LGR5-expressing cells are myoepithelial cells that are positive for P63 (indicated by black arrows). Scale bar: 50 μm.
LGR5 expression in four subtypes of invasive breast cancers
We measured LGR5 H-scores in a large cohort of invasive BC (n = 279) and a total of 18 cases of LGR5-expressing BC were observed. The pathological features of them are shown in Table 1, and 9 cases with an H-score of 10 or higher were considered positive for statistical analysis. Interestingly, in some cases, we observed a remarkable increase in LGR5 expression in the myoepithelium of nonneoplastic ducts surrounded by cancer cells (Supplementary Fig. S2). The associations between LGR5 positivity and clinicopathological characteristics are summarized in Table 2. Histologically, LGR5 expression was only observed in invasive carcinomas of no special type. It was associated with poor tubule formation, marked nuclear pleomorphism (P < 0.001), and a high mitotic count (P < 0.001). Thus, it was not surprising to find that all LGR5-positive BC were scored as grade 3 (P < 0.001). LGR5 expression was more frequently observed in BC with higher T stages (P < 0.001), whereas there were no correlations with lymphovascular invasion (P = 0.428), N stages (P = 0.748), or AJCC (7th edition) tumor stages (P = 0.545). Based on ER, PR, and HER2 positivity, BCs were classified into four molecular subtypes: luminal A, luminal B, HER2, and TNBC. Interestingly, LGR5 positivity showed strong negative correlations with ER (P < 0.001) and PR (P < 0.001) expressions, and it turned out that all LGR5-positive BCs except one belonged to the TNBC subtype (P < 0.001), comprising 21% of all TNBCs (8 out of 38 cases). Representative images, including H&E stain, immunohistochemical stain for ER, PR, and HER2, and in situ hybridization for LGR5 are shown in Fig. 3A. When comparing LGR5 H-scores between molecular subtypes, they were significantly higher in TNBC than in other types (Fig. 3B). As LGR5 is one of the Wnt target genes, we additionally explored whether Wnt/b-catenin signaling activity is responsible for LGR5 expression in TNBC by evaluating the nuclear expression of b-catenin, indicative of upregulated Wnt signaling. However, none of the LGR5-positive BCs showed nuclear b-catenin positivity (Supplementary Fig. S3).
LGR5 Expression in Invasive Breast Cancers of Four Molecular Subtypes. (A) Representative H&E staining, immunohistochemical staining (ER, PR, and HER2), and in situ hybridization (LGR5) according to four subtypes of breast cancers. (B) A graph showing histoscores of LGR5 in BCs. ER, estrogen receptor; PR, progesterone receptor; TNBC, triple negative breast cancer. Ns, not significant. ***P < 0.001. Scale bars: 100 μm.
Associations of LGR5 with EGFR, CK5/6, Ki-67, and P53
The TNBC subgroup was first revealed by microarray-based expression profiling studies25. They are known to have particular pathological and molecular characteristics besides the lack of ER, PR, and HER2 expression: high histologic grade, high Ki-67 index, occasional presence of medullary or metaplastic elements, positivity for EGFR, CK5/6, and frequent TP53 mutations26,27. As the vast majority of LGR5-positive BCs belong to TNBC, we investigated whether there are any correlations between LGR5 expression levels and those distinct TNBC features. Representative images of an LGR5-positive BC showing high levels of EGFR, CK5/6, and Ki-67, as well as a complete loss of P53, are presented in Fig. 4A. We measured the EGFR H-scores and CK5/6 expression, as well as the percentages of Ki-67- and P53-positive cancer cells in 18 LGR5-expressing BCs. However, when evaluating their correlations to LGR5 H-scores, none of them exhibited significant associations (Fig. 4B).
LGR5 Expression in Triple Negative Breast Cancers. (A) Representative images of LGR5 expression of a triple negative breast cancer, showing high expression for EGFR, CK5/6, and Ki-67, but negativity for P53. (B) Scatter plots showing the correlations of LGR5 H-score with EGFR, CK5/6, Ki-67, and P53 expression in LGR5-positive BCs (n = 18). Scale bars: 100 μm.
Discussion
Using RNA in situ hybridization, we thoroughly investigated LGR5 expression in normal, benign, and malignant human breast tumors. Our study demonstrated that unlike the murine model, LGR5-positive cells are not present in the proximal or distal ducts of adult mammary tissues. However, this can possibly be because of the limitation of our detection method. In the mouse model, lineage tracing has been used to visualize LGR5 expression using a fluorescent reporter protein, whereas here we used ISH to determine the expression of LGR5 mRNA. It may be that current RNA ISH techniques are not yet sensitive enough to detect cells with very low levels of LGR5 that might exist in human mammary tissues. Additionally, as we had obtained all breast samples examined in this study from adult patients, it remains to be evaluated whether developmental stage, hormone status during menstruation, or pregnancy have any influence on LGR5 expression.
In mice, LGR5 is expressed in 2% to 3% of mammary epithelial cells and localized to the nipple region, and the vast majority of LGR5+ cells are myoepithelial cells7,22. Fu et al. have suggested distinct mammary stem cell subsets, proximally restricted LGR5+/Tspan8hi cells in a deeply quiescent state can be activated by ovarian hormones and a separate pool of LGR5+/Tspan8- cells in the distal portion of mammary trees24. In this study, we did not find any evidence of LGR5-positive resident stem cells in the human breast. Instead, LGR5-positive myoepithelial cells were observed in the scar caused by previous needle biopsies and in an inflamed tissue area formed by excision. This finding is consistent with a previous report showing that LGR5+ cells are efficient in reconstituting murine mammary glands7. In addition, a similar expression pattern of LGR5 has been most recently reported in the skeletal muscle regeneration. Leung et al. have shown that LGR5 is not expressed in the satellite cells of uninjured muscle, however, it is upregulated in myogenic progenitor cells after skeletal muscle injury and LGR5+ cells contribute to muscle regeneration and satellite cell pool replenishment28. Therefore, it seems that in human mammary tissues, LGR5 cells are recruited to function as facultative stem cells responsible for tissue renewal following injury. Further study is required to confirm that LGR5 cells are a response to stem cell population to tissue damage in the human breast. An example of this is using the in vitro breast organoid system to investigate LGR5 expression during regeneration following epithelial cell damage.
In contrast to the absence of LGR5 expression in normal mammary tissues, it is surprising to find that LGR5 cells are frequently detected in DCIS attenuated basal myoepithelial cells. (Fig. 1B and Supplementary Fig. S1). The mammary myoepithelial cells are involved in mammary gland development and normally facilitate milk expulsion during lactation. Studies suggest that myoepithelial cells play a tumor suppressive function by secreting various proteins such as maspin, p63, Wilms tumor 1, and laminin 129,30,31. With DCIS progression, myoepithelial cells surrounding them become flat and are gradually lost, resulting in the transition from preinvasive to invasive cancer32. Therefore, it is reasonable to speculate that the appearance of LGR5-positive cells in the DCIS myoepithelium can be attributed to them sensing the pressure of an increasing number of cancer cells as a signal of tissue injury. Likewise, we also observed a dramatic increase in the number of LGR5 cells in the myoepithelium of nontumorous ducts entrapped by invasive cancer cells (Supplementary Fig. S2). Alternatively, the induced LGR5 expression in the myoepithelium might be the consequence of intricate interactions between the myoepithelium and cancer cells.
In a large cohort, we found that a total 3% of invasive BC are positive for LGR5. Comparing this to our previous studies using the same RNA ISH technique, the positivity is similar to that in gastric cancers (7%)33 and much lower than that in colorectal cancers (68%)34. Overall LGR5 cancer positivity is generally associated with the basal LGR5 expression levels in each organ, as the stomach and breast show very little or no LGR5 expression in normal tissues, whereas the colorectum has a greater number of LGR5 cells at the base of the crypts. Considering that LGR5 cells are the origin of cancers in the stomach10,12 and colorectum11, its low positivity in BC is probably because of the scarcity of LGR5 cells in the homeostatic state. More interestingly, LGR5 positivity was exclusively observed in the TNBC subtype. Although there was one luminal A-BC that was positive for LGR5, its Allred score for ER was 3, and it showed high levels of CK5/6 expression, suggesting that it also harbors TNBC features. This is consistent with the previous finding from the analysis of BC samples in METABRIC database, where LGR5 mRNA expression is significantly higher in TNBC as compared to luminal A, luminal B, and HER2 subtypes19. Because the expression of LGR5 was observed exclusively in basal myoepithelial cells under regenerative conditions, it is probable that those reappearing LGR5 cells might represent the cells of BC’s origin of the TNBC subtype.
TNBC that accounts for 10% to 20% of all BC is a highly diverse group simply defined by the absence of ER/PR/HER-2. For better molecular-based targeted therapies, there have been efforts to identify the subtypes in TNBC. For instance, Lehmann et al. suggested six different molecular subtypes of TNBC through genomic-wide gene expression profiling analyses35. More recently, four stable TNBC subtypes characterized by the expression of molecular profiles with distinct prognoses have been described by Burstein et al.: luminal androgen receptor, mesenchymal, basal-like immunosuppressed, and basal-like immune-activated (BLIA).36. Even though LGR5 was not identified as one of the biomarkers that define subgroups in the above-mentioned studies, it would be interesting to investigate to which subtype LGR5-positive TNBC belongs. This would contribute to a better understanding of the molecular characteristics of LGR5-positive BC.
LGR5-positive cells have been shown to be the cancer stem cells responsible for tumor growth and metastasis in CRCs15,16. Yang et al. have suggested that in BC, LGR5 + cells promote cancer cell mobility, tumor formation, epithelial-mesenchymal transition, as well as stemness by activating Wnt signaling17. More recently, Hagerling et al. showed a role for LGR5 in tumor initiation in TNBC through different lineage-tracing experiments that revealed a therapeutic potential of anti-LGR5 to target LGR5 + cells in an aggressive ER-negative BC19. Although we did not continue to investigate the functional implication of LGR5 in TNBC, we discovered an absence of LGR5 + cells in the normal mammary tissues and specific LGR5 expression in TNBC subtypes. These findings suggest that they would be less likely to have side effects on normal breast tissue while anti-LGR5 therapy exerts its effects on cancer cells.
As one of the Wnt target genes, LGR5 expression has been associated with abnormally enhanced Wnt signaling in many different types of cancers. We previously showed the positive correlations between LGR5 and nuclear ß-catenin expression in gastric33 and colorectal cancers34. For BC, nuclear ß-catenin was reported mostly in TNBC, although CTNNB1 mutations were not identified37,38, suggesting the implication of Wnt pathway activation in TNBC. However, no TNBC identified in this study displayed nuclear ß-catenin expression. Representative images are shown in Supplementary Fig. S3. This discrepancy might be because of the small number of TNBC cases in our study or in the differences in the BC patient cohort. Different criteria for nuclear ß-catenin positivity between studies might have resulted in contrary results. Further study is needed to find out whether activated Wnt signaling is involved in LGR5 expression in TNBC or if signaling pathways other than canonical Wnt signaling are responsible for LGR5 induction.
In summary, LGR5 cells are not normally found in the adult human breast. However, they appear in regenerative conditions such as tissue injury, degeneration by DCIS, or entrapment by cancer cells in the mammary duct myoepithelium. This myoepithelium-restricted LGR5 expression may be related to the specific and frequent LGR5 expression in invasive BC of the TNBC subtype. Further studies on the functional significance of LGR5 are required to explore LGR5 as a potential therapeutic target for LGR5-positive TNBC.
References
Park, S. et al. Unlike LGR4, LGR5 potentiates Wnt–β-catenin signaling without sequestering E3 ligases. Sci. Signal. 13, 4051 (2020).
de Lau, W., Peng, W. C., Gros, P. & Clevers, H. The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength. Genes Dev. 28, 305–316 (2014).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Jaks, V. et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 40, 1291 (2008).
Barker, N. et al. Lgr5+ ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010).
Ng, A. et al. Lgr5 marks stem/progenitor cells in ovary and tubal epithelia. Nat. Cell Biol. 16, 745–757 (2014).
Plaks, V. et al. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 3, 70–78 (2013).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013).
Leushacke, M. et al. Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat. Cell Biol. 19, 774–786 (2017).
Barker, N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 (2009).
Li, X.-B. et al. Gastric Lgr5+ stem cells are the cellular origin of invasive intestinal-type gastric cancer in mice. Cell Res. 26, 838–849 (2016).
Huang, P. Y. et al. Lgr6 is a stem cell marker in mouse skin squamous cell carcinoma. Nat. Genet. 49, 1624 (2017).
Varga, J. & Greten, F. R. Cell plasticity in epithelial homeostasis and tumorigenesis. Nat. Cell Biol. 19, 1133–1141 (2017).
Shimokawa, M. et al. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 545, 187–192 (2017).
e Melo, F. D. et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680 (2017).
Yang, L. et al. LGR5 promotes breast cancer progression and maintains stem-like cells through activation of W nt/β-catenin signaling. Stem Cells 33, 2913–2924 (2015).
Hou, M.-F., Chen, P.-M. & Chu, P.-Y. LGR5 overexpression confers poor relapse-free survival in breast cancer patients. BMC Cancer 18, 219 (2018).
Hagerling, C. et al. LGR5 in breast cancer and ductal carcinoma in situ: A diagnostic and prognostic biomarker and a therapeutic target. BMC Cancer 20, 1–14 (2020).
Ogasawara, S. et al. Correlation of clinicopathological features and LRG5 expression in triple-negative breast carcinoma. Ann. Diagn. Pathol. 46, 151491 (2020).
Wolff, A. C. et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch. Pathol. Lab. Med. 131, 18–43 (2007).
Van Keymeulen, A. et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189–193 (2011).
Wang, D. et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature 517, 81–84 (2015).
Fu, N. Y. et al. Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat. Cell Biol. 19, 164–176 (2017).
Perou, C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
Foulkes, W. D., Smith, I. E. & Reis-Filho, J. S. Triple-negative breast cancer. N. Engl. J. Med. 363, 1938–1948 (2010).
Aleskandarany, M. A. et al. Prognostic value of proliferation assay in the luminal, HER2-positive, and triple-negative biologic classes of breast cancer. Breast Cancer Res. 14, 1–11 (2012).
Leung, C. et al. Lgr5 marks adult progenitor cells contributing to skeletal muscle regeneration and sarcoma formation. Cell Rep 33, 108535 (2020).
Zou, Z. et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science 263, 526–529 (1994).
Barbareschi, M. et al. p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am. J. Surg. Pathol. 25, 1054–1060 (2001).
Li, J. H. & Man, Y. G. Dual usages of single Wilms’ tumor 1 immunohistochemistry in evaluation of breast tumors: A preliminary study of 30 cases 1. Cancer Biomark. 5, 109–116 (2009).
Russell, T. D. et al. Myoepithelial cell differentiation markers in ductal carcinoma in situ progression. Am. J. Pathol. 185, 3076–3089 (2015).
Jang, B. G., Lee, B. L. & Kim, W. H. Prognostic significance of leucine-rich-repeat-containing G-protein-coupled receptor 5, an intestinal stem cell marker, in gastric carcinomas. Gastric Cancer 19, 767–777 (2016).
Jang, B. G. et al. Expression profile of LGR5 and its prognostic significance in colorectal cancer progression. Am. J. Pathol. 188, 2236–2250 (2018).
Lehmann, B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 121, 2750–2767 (2011).
Burstein, M. D. et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 21, 1688–1698 (2015).
Khramtsov, A. I. et al. Wnt/β-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am. J. Pathol. 176, 2911–2920 (2010).
Geyer, F. C. et al. β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod. Pathol. 24, 209–231 (2011).
Acknowledgements
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (No.2019R1F1A1059250) (to B.J.), (No.2019M3E5D1A01069361) (to S.J. L.), (No. 2020R1I1A1A01069168) (to H.S.K.) and National Research Foundation of Korea (NRF) Basic Science Research Program, Ministry of Science, ICT & Future Planning grants (NRF-2017R1C1B5075941) (to H.J.L) and the Soonchunhyang University Research Fund (to H.J.L.).
Author information
Authors and Affiliations
Contributions
H.J.L. and J.K.M. performed the experiments and drafted the manuscript. J.H.C. contributed to collection of breast tissues. H.S.K., H.S.G., H.M.K., D.H.K. and S.J.L. contributed to data collection and construction of TMAs. B.J. designed the study, supervised the experiments, interpreted the results and revised the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, H.J., Myung, J.K., Kim, H.S. et al. Expression of LGR5 in mammary myoepithelial cells and in triple-negative breast cancers. Sci Rep 11, 17750 (2021). https://doi.org/10.1038/s41598-021-97351-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-97351-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.